Note
Click here to download the full example code
Reading and writing an evoked file¶
This script shows how to read and write evoked datasets.
# Author: Alexandre Gramfort <alexandre.gramfort@inria.fr>
#
# License: BSD (3-clause)
from mne import read_evokeds
from mne.datasets import sample
print(__doc__)
data_path = sample.data_path()
fname = data_path + '/MEG/sample/sample_audvis-ave.fif'
# Reading
condition = 'Left Auditory'
evoked = read_evokeds(fname, condition=condition, baseline=(None, 0),
proj=True)
Out:
Reading /home/circleci/mne_data/MNE-sample-data/MEG/sample/sample_audvis-ave.fif ...
Read a total of 4 projection items:
PCA-v1 (1 x 102) active
PCA-v2 (1 x 102) active
PCA-v3 (1 x 102) active
Average EEG reference (1 x 60) active
Found the data of interest:
t = -199.80 ... 499.49 ms (Left Auditory)
0 CTF compensation matrices available
nave = 55 - aspect type = 100
Projections have already been applied. Setting proj attribute to True.
Applying baseline correction (mode: mean)
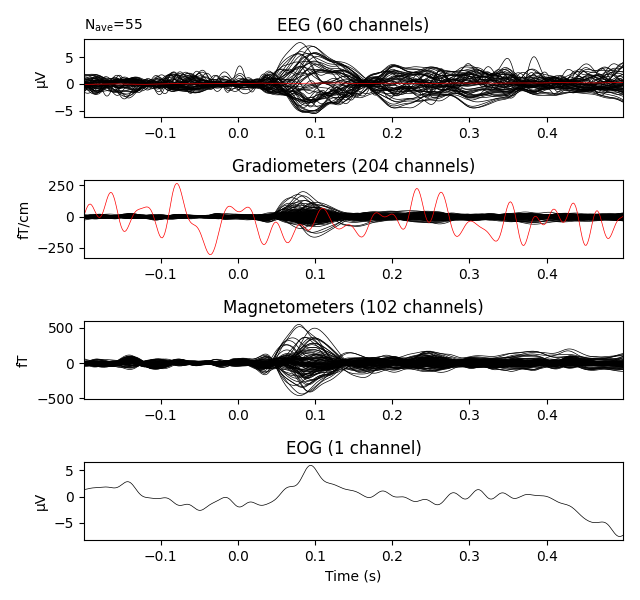
Show result as a butterfly plot: By using exclude=[] bad channels are not excluded and are shown in red
evoked.plot(exclude=[], time_unit='s')
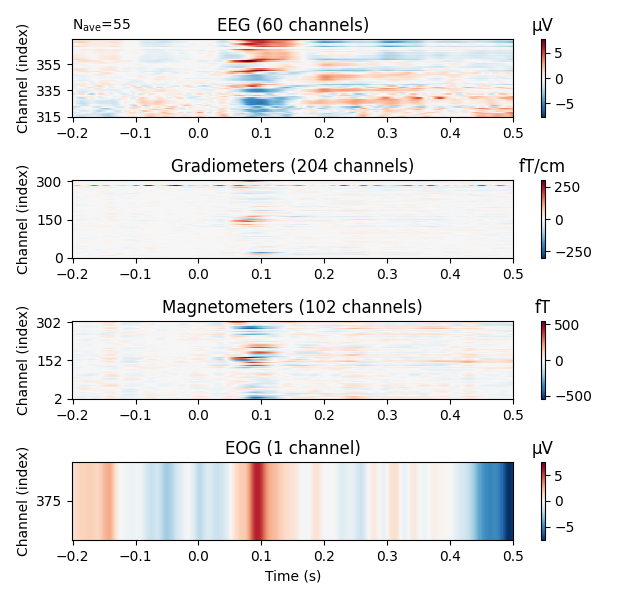
# Show result as a 2D image (x: time, y: channels, color: amplitude)
evoked.plot_image(exclude=[], time_unit='s')
Use mne.Evoked.save() or mne.write_evokeds() to write the evoked
responses to a file.
Total running time of the script: ( 0 minutes 2.825 seconds)
Estimated memory usage: 10 MB