mne_bids.get_head_mri_trans#
- mne_bids.get_head_mri_trans(bids_path, extra_params=None, t1_bids_path=None, fs_subject=None, fs_subjects_dir=None, *, kind=None, verbose=None)[source]#
Produce transformation matrix from MEG and MRI landmark points.
Will attempt to read the landmarks of Nasion, LPA, and RPA from the sidecar files of (i) the MEG and (ii) the T1-weighted MRI data. The two sets of points will then be used to calculate a transformation matrix from head coordinates to MRI coordinates.
Note
The MEG and MRI data need not necessarily be stored in the same session or even in the same BIDS dataset. See the
t1_bids_pathparameter for details.- Parameters:
- bids_path
BIDSPath The path of the electrophysiology recording. If
datatypeandsuffixare not present, they will be set to'meg', and a warning will be raised.Changed in version 0.10: A warning is raised it
datatypeorsuffixare not set.- extra_params
None|dict Extra parameters to be passed to
mne.io.read_raw()when reading the MEG file.- t1_bids_path
BIDSPath|None If
None(default), will try to discover the T1-weighted MRI file based on the name and location of the MEG recording specified via thebids_pathparameter. Alternatively, you explicitly specify which T1-weighted MRI scan to use for extraction of MRI landmarks. To do that, pass amne_bids.BIDSPathpointing to the scan. Use this parameter e.g. if the T1 scan was recorded during a different session than the MEG. It is even possible to point to a T1 image stored in an entirely different BIDS dataset than the MEG data.- fs_subject
str The subject identifier used for FreeSurfer.
Changed in version 0.10: Does not default anymore to
bids_path.subjectifNone.- fs_subjects_dirpath-like |
None The FreeSurfer subjects directory. If
None, defaults to theSUBJECTS_DIRenvironment variable.Added in version 0.8.
- kind
str|None The suffix of the anatomical landmark names in the JSON sidecar. A suffix might be present e.g. to distinguish landmarks between sessions. If provided, should not include a leading underscore
_. For example, if the landmark names in the JSON sidecar file areLPA_ses-1,RPA_ses-1,NAS_ses-1, you should pass'ses-1'here. IfNone, no suffix is appended, the landmarks namedNasion(orNAS),LPA, andRPAwill be used.Added in version 0.10.
- verbose
bool|str|int|None Control verbosity of the logging output. If
None, use the default verbosity level. See the logging documentation andmne.verbose()for details. Should only be passed as a keyword argument.
- bids_path
- Returns:
- trans
mne.transforms.Transform The data transformation matrix from head to MRI coordinates.
- trans
Examples using mne_bids.get_head_mri_trans#
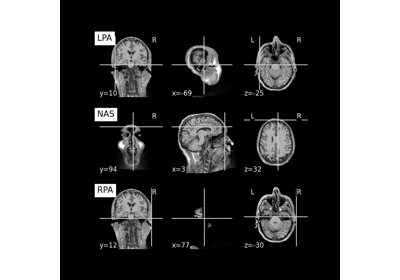
07. Save and load T1-weighted MRI scan along with anatomical landmarks in BIDS