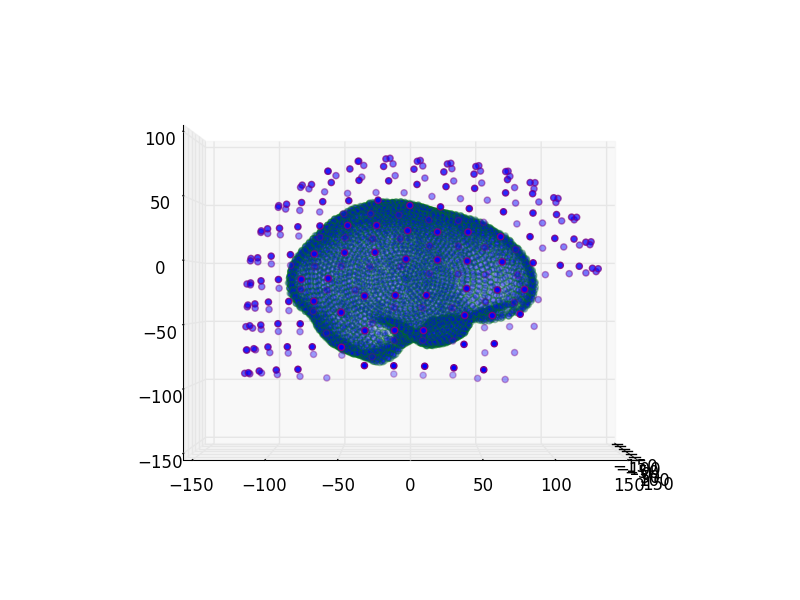
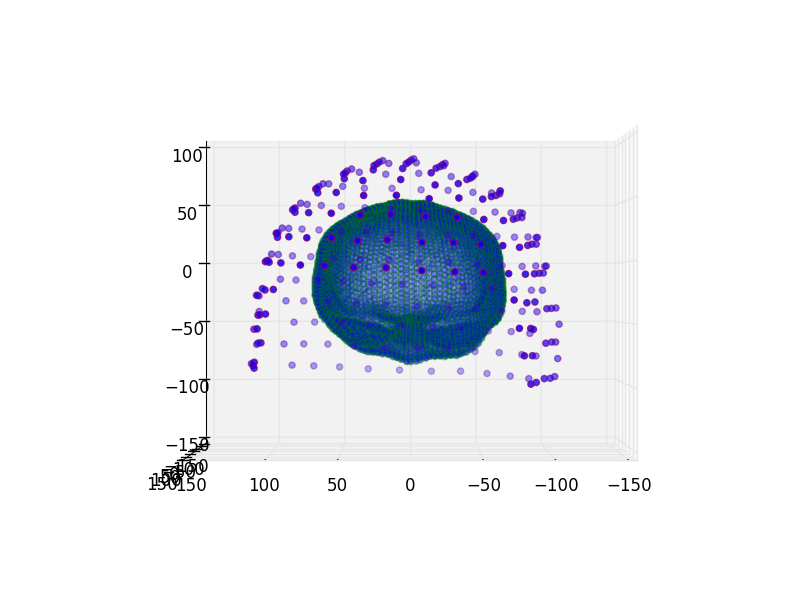
Read and plot coregistration¶
We’ll take a look at the coregistration here.
# Author: Denis A. Enegemann
# License: BSD 3 clause
import os.path as op
from hcp.viz import plot_coregistration
we assume our data is inside a designated folder under $HOME
storage_dir = op.expanduser('~/mne-hcp-data')
and we assume to have the downloaded data, the MNE/freesurfer style
anatomy directory, and the MNE style MEG directory.
these can be obtained from make_mne_anatomy().
See also Extract essential anatomy information.
hcp_params = dict(
subject='105923',
hcp_path=op.join(storage_dir, 'HCP'),
subjects_dir=op.join(storage_dir, 'subjects'),
recordings_path=op.join(storage_dir, 'hcp-meg'))
let’s plot two views
for azim in (0, 90):
plot_coregistration(
view_init=(('azim', azim), ('elev', 0)), **hcp_params)
Out:
Reading 4D PDF file /Users/dengemann/mne-hcp-data/HCP/105923/unprocessed/MEG/3-Restin/4D/c,rfDC...
Creating Neuromag info structure ...
... Setting channel info structure.
... no headshape file supplied, doing nothing.
Currently direct inclusion of 4D weight tables is not supported. For critical use cases please take into account the MNE command "mne_create_comp_data" to include weights as printed out by the 4D "print_table" routine.
Triangle file: nvert = 5000 ntri = 9996
Reading 4D PDF file /Users/dengemann/mne-hcp-data/HCP/105923/unprocessed/MEG/3-Restin/4D/c,rfDC...
Creating Neuromag info structure ...
... Setting channel info structure.
... no headshape file supplied, doing nothing.
Currently direct inclusion of 4D weight tables is not supported. For critical use cases please take into account the MNE command "mne_create_comp_data" to include weights as printed out by the 4D "print_table" routine.
Triangle file: nvert = 5000 ntri = 9996
Total running time of the script: ( 0 minutes 0.451 seconds)