Note
Click here to download the full example code
Compute envelope correlations in source space¶
Compute envelope correlations of orthogonalized activity 1 2 in source space using resting state CTF data.
# Authors: Eric Larson <larson.eric.d@gmail.com>
# Sheraz Khan <sheraz@khansheraz.com>
# Denis Engemann <denis.engemann@gmail.com>
#
# License: BSD (3-clause)
import os.path as op
import numpy as np
import matplotlib.pyplot as plt
import mne
from mne.connectivity import envelope_correlation
from mne.minimum_norm import make_inverse_operator, apply_inverse_epochs
from mne.preprocessing import compute_proj_ecg, compute_proj_eog
data_path = mne.datasets.brainstorm.bst_resting.data_path()
subjects_dir = op.join(data_path, 'subjects')
subject = 'bst_resting'
trans = op.join(data_path, 'MEG', 'bst_resting', 'bst_resting-trans.fif')
src = op.join(subjects_dir, subject, 'bem', subject + '-oct-6-src.fif')
bem = op.join(subjects_dir, subject, 'bem', subject + '-5120-bem-sol.fif')
raw_fname = op.join(data_path, 'MEG', 'bst_resting',
'subj002_spontaneous_20111102_01_AUX.ds')
Here we do some things in the name of speed, such as crop (which will hurt SNR) and downsample. Then we compute SSP projectors and apply them.
raw = mne.io.read_raw_ctf(raw_fname, verbose='error')
raw.crop(0, 60).pick_types(meg=True, eeg=False).load_data().resample(80)
raw.apply_gradient_compensation(3)
projs_ecg, _ = compute_proj_ecg(raw, n_grad=1, n_mag=2)
projs_eog, _ = compute_proj_eog(raw, n_grad=1, n_mag=2, ch_name='MLT31-4407')
raw.info['projs'] += projs_ecg
raw.info['projs'] += projs_eog
raw.apply_proj()
cov = mne.compute_raw_covariance(raw) # compute before band-pass of interest
Out:
Including 0 SSP projectors from raw file
Running ECG SSP computation
Reconstructing ECG signal from Magnetometers
Setting up band-pass filter from 5 - 35 Hz
FIR filter parameters
---------------------
Designing a two-pass forward and reverse, zero-phase, non-causal bandpass filter:
- Windowed frequency-domain design (firwin2) method
- Hann window
- Lower passband edge: 5.00
- Lower transition bandwidth: 0.50 Hz (-12 dB cutoff frequency: 4.75 Hz)
- Upper passband edge: 35.00 Hz
- Upper transition bandwidth: 0.50 Hz (-12 dB cutoff frequency: 35.25 Hz)
- Filter length: 800 samples (10.000 sec)
Number of ECG events detected : 88 (average pulse 88 / min.)
Computing projector
Not setting metadata
Not setting metadata
88 matching events found
No baseline correction applied
0 projection items activated
Loading data for 88 events and 49 original time points ...
Rejecting epoch based on MAG : ['MLT31-4407', 'MRT31-4407']
Rejecting epoch based on MAG : ['MLT31-4407', 'MLT41-4407', 'MRT31-4407', 'MRT41-4407']
Rejecting epoch based on MAG : ['MLT31-4407', 'MLT41-4407', 'MRT31-4407', 'MRT41-4407']
4 bad epochs dropped
No gradiometers found. Forcing n_grad to 0
No EEG channels found. Forcing n_eeg to 0
Adding projection: axial--0.200-0.400-PCA-01
Adding projection: axial--0.200-0.400-PCA-02
Done.
Including 0 SSP projectors from raw file
Running EOG SSP computation
Using channel MLT31-4407 as EOG channel
EOG channel index for this subject is: [137]
Filtering the data to remove DC offset to help distinguish blinks from saccades
Setting up band-pass filter from 1 - 10 Hz
FIR filter parameters
---------------------
Designing a two-pass forward and reverse, zero-phase, non-causal bandpass filter:
- Windowed frequency-domain design (firwin2) method
- Hann window
- Lower passband edge: 1.00
- Lower transition bandwidth: 0.50 Hz (-12 dB cutoff frequency: 0.75 Hz)
- Upper passband edge: 10.00 Hz
- Upper transition bandwidth: 0.50 Hz (-12 dB cutoff frequency: 10.25 Hz)
- Filter length: 800 samples (10.000 sec)
Now detecting blinks and generating corresponding events
Found 12 significant peaks
Number of EOG events detected : 12
Computing projector
Not setting metadata
Not setting metadata
12 matching events found
No baseline correction applied
0 projection items activated
Loading data for 12 events and 33 original time points ...
Rejecting epoch based on MAG : ['MRT41-4407']
1 bad epochs dropped
No gradiometers found. Forcing n_grad to 0
No EEG channels found. Forcing n_eeg to 0
Adding projection: axial--0.200-0.200-PCA-01
Adding projection: axial--0.200-0.200-PCA-02
Done.
Using up to 300 segments
Number of samples used : 4800
[done]
Now we band-pass filter our data and create epochs.
raw.filter(14, 30)
events = mne.make_fixed_length_events(raw, duration=5.)
epochs = mne.Epochs(raw, events=events, tmin=0, tmax=5.,
baseline=None, reject=dict(mag=8e-13), preload=True)
del raw
Out:
Not setting metadata
Not setting metadata
12 matching events found
No baseline correction applied
Created an SSP operator (subspace dimension = 4)
4 projection items activated
Loading data for 12 events and 401 original time points ...
Rejecting epoch based on MAG : ['MRC42-4407', 'MRC54-4407', 'MRP12-4407', 'MRP22-4407', 'MRP23-4407']
2 bad epochs dropped
Compute the forward and inverse¶
src = mne.read_source_spaces(src)
fwd = mne.make_forward_solution(epochs.info, trans, src, bem)
inv = make_inverse_operator(epochs.info, fwd, cov)
del fwd, src
Out:
Reading a source space...
Computing patch statistics...
Patch information added...
Distance information added...
[done]
Reading a source space...
Computing patch statistics...
Patch information added...
Distance information added...
[done]
2 source spaces read
Source space : <SourceSpaces: [<surface (lh), n_vertices=156869, n_used=4098>, <surface (rh), n_vertices=155654, n_used=4098>] MRI (surface RAS) coords, subject 'bst_resting', ~27.4 MB>
MRI -> head transform : /home/circleci/mne_data/MNE-brainstorm-data/bst_resting/MEG/bst_resting/bst_resting-trans.fif
Measurement data : instance of Info
Conductor model : /home/circleci/mne_data/MNE-brainstorm-data/bst_resting/subjects/bst_resting/bem/bst_resting-5120-bem-sol.fif
Accurate field computations
Do computations in head coordinates
Free source orientations
Read 2 source spaces a total of 8196 active source locations
Coordinate transformation: MRI (surface RAS) -> head
0.999797 -0.005775 -0.019288 2.71 mm
0.011390 0.952195 0.305279 16.66 mm
0.016602 -0.305437 0.952068 28.47 mm
0.000000 0.000000 0.000000 1.00
Read 272 MEG channels from info
Read 26 MEG compensation channels from info
99 coil definitions read
Coordinate transformation: MEG device -> head
0.998490 -0.050225 -0.022235 1.90 mm
0.052235 0.993447 0.101656 13.13 mm
0.016984 -0.102664 0.994571 66.69 mm
0.000000 0.000000 0.000000 1.00
5 compensation data sets in info
MEG coil definitions created in head coordinates.
Removing 5 compensators from info because not all compensation channels were picked.
Source spaces are now in head coordinates.
Setting up the BEM model using /home/circleci/mne_data/MNE-brainstorm-data/bst_resting/subjects/bst_resting/bem/bst_resting-5120-bem-sol.fif...
Loading surfaces...
Loading the solution matrix...
Homogeneous model surface loaded.
Loaded linear_collocation BEM solution from /home/circleci/mne_data/MNE-brainstorm-data/bst_resting/subjects/bst_resting/bem/bst_resting-5120-bem-sol.fif
Employing the head->MRI coordinate transform with the BEM model.
BEM model bst_resting-5120-bem-sol.fif is now set up
Source spaces are in head coordinates.
Checking that the sources are inside the surface (will take a few...)
Skipping interior check for 1239 sources that fit inside a sphere of radius 49.6 mm
Skipping solid angle check for 0 points using Qhull
Skipping interior check for 1235 sources that fit inside a sphere of radius 49.6 mm
Skipping solid angle check for 0 points using Qhull
Setting up compensation data...
272 out of 272 channels have the compensation set.
Desired compensation data (3) found.
All compensation channels found.
Preselector created.
Compensation data matrix created.
Postselector created.
Composing the field computation matrix...
Composing the field computation matrix (compensation coils)...
Computing MEG at 8196 source locations (free orientations)...
Finished.
Converting forward solution to surface orientation
Average patch normals will be employed in the rotation to the local surface coordinates....
Converting to surface-based source orientations...
[done]
Computing inverse operator with 272 channels.
272 out of 272 channels remain after picking
Removing 5 compensators from info because not all compensation channels were picked.
Selected 272 channels
Creating the depth weighting matrix...
272 magnetometer or axial gradiometer channels
limit = 8074/8196 = 10.006553
scale = 5.92189e-11 exp = 0.8
Applying loose dipole orientations to surface source spaces: 0.2
Whitening the forward solution.
Removing 5 compensators from info because not all compensation channels were picked.
Created an SSP operator (subspace dimension = 4)
Computing rank from covariance with rank=None
Using tolerance 5.5e-14 (2.2e-16 eps * 272 dim * 0.92 max singular value)
Estimated rank (mag): 268
MAG: rank 268 computed from 272 data channels with 4 projectors
Setting small MAG eigenvalues to zero (without PCA)
Creating the source covariance matrix
Adjusting source covariance matrix.
Computing SVD of whitened and weighted lead field matrix.
largest singular value = 3.42374
scaling factor to adjust the trace = 1.2238e+19
Compute label time series and do envelope correlation¶
labels = mne.read_labels_from_annot(subject, 'aparc_sub',
subjects_dir=subjects_dir)
epochs.apply_hilbert() # faster to apply in sensor space
stcs = apply_inverse_epochs(epochs, inv, lambda2=1. / 9., pick_ori='normal',
return_generator=True)
label_ts = mne.extract_label_time_course(
stcs, labels, inv['src'], return_generator=True)
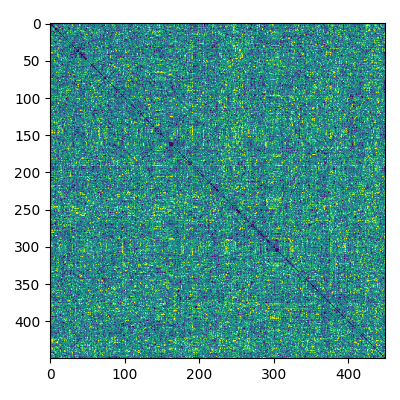
corr = envelope_correlation(label_ts, verbose=True)
# let's plot this matrix
fig, ax = plt.subplots(figsize=(4, 4))
ax.imshow(corr, cmap='viridis', clim=np.percentile(corr, [5, 95]))
fig.tight_layout()
Out:
Reading labels from parcellation...
read 226 labels from /home/circleci/mne_data/MNE-brainstorm-data/bst_resting/subjects/bst_resting/label/lh.aparc_sub.annot
read 224 labels from /home/circleci/mne_data/MNE-brainstorm-data/bst_resting/subjects/bst_resting/label/rh.aparc_sub.annot
Preparing the inverse operator for use...
Scaled noise and source covariance from nave = 1 to nave = 1
Created the regularized inverter
Created an SSP operator (subspace dimension = 4)
Created the whitener using a noise covariance matrix with rank 268 (4 small eigenvalues omitted)
Computing noise-normalization factors (dSPM)...
[done]
Picked 272 channels from the data
Computing inverse...
Eigenleads need to be weighted ...
Processing epoch : 1 / 10
Extracting time courses for 450 labels (mode: mean_flip)
Processing epoch : 2 / 10
Extracting time courses for 450 labels (mode: mean_flip)
Processing epoch : 3 / 10
Extracting time courses for 450 labels (mode: mean_flip)
Processing epoch : 4 / 10
Extracting time courses for 450 labels (mode: mean_flip)
Processing epoch : 5 / 10
Extracting time courses for 450 labels (mode: mean_flip)
Processing epoch : 6 / 10
Extracting time courses for 450 labels (mode: mean_flip)
Processing epoch : 7 / 10
Extracting time courses for 450 labels (mode: mean_flip)
Processing epoch : 8 / 10
Extracting time courses for 450 labels (mode: mean_flip)
Processing epoch : 9 / 10
Extracting time courses for 450 labels (mode: mean_flip)
Processing epoch : 10 / 10
Extracting time courses for 450 labels (mode: mean_flip)
[done]
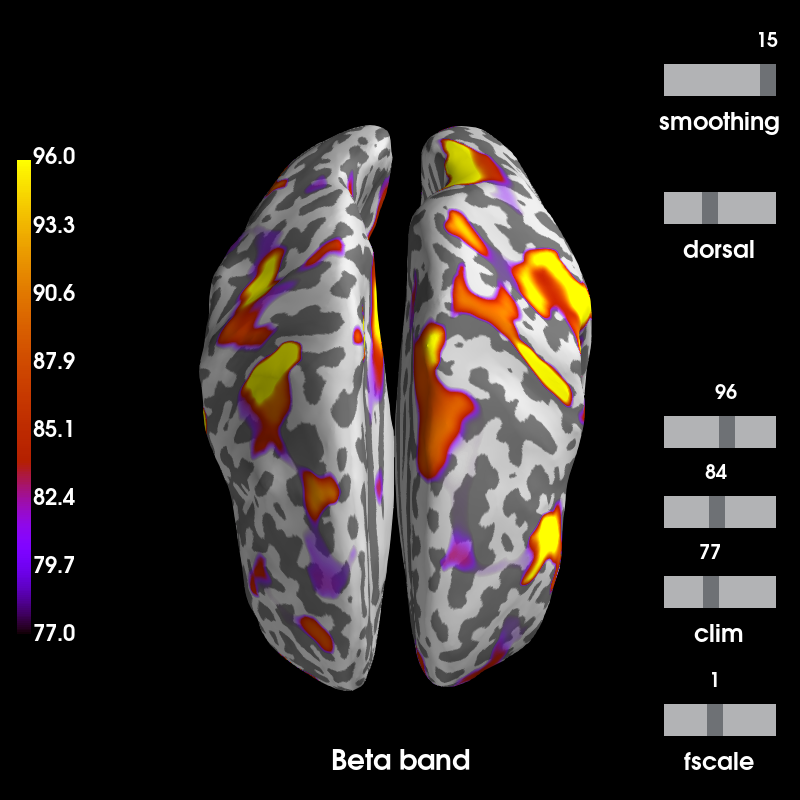
Compute the degree and plot it¶
threshold_prop = 0.15 # percentage of strongest edges to keep in the graph
degree = mne.connectivity.degree(corr, threshold_prop=threshold_prop)
stc = mne.labels_to_stc(labels, degree)
stc = stc.in_label(mne.Label(inv['src'][0]['vertno'], hemi='lh') +
mne.Label(inv['src'][1]['vertno'], hemi='rh'))
brain = stc.plot(
clim=dict(kind='percent', lims=[75, 85, 95]), colormap='gnuplot',
subjects_dir=subjects_dir, views='dorsal', hemi='both',
smoothing_steps=25, time_label='Beta band')
Out:
Using control points [77. 84. 96.]
References¶
- 1
Hipp JF, Hawellek DJ, Corbetta M, Siegel M, Engel AK (2012) Large-scale cortical correlation structure of spontaneous oscillatory activity. Nature Neuroscience 15:884–890
- 2
Khan S et al. (2018). Maturation trajectories of cortical resting-state networks depend on the mediating frequency band. Neuroimage 174:57–68
Total running time of the script: ( 0 minutes 42.176 seconds)
Estimated memory usage: 845 MB