Note
Click here to download the full example code
Querying epochs with rich metadata¶
Selecting a subset of epochs based on rich metadata.
MNE allows you to include metadata along with your mne.Epochs objects.
This is in the form of a pandas.DataFrame that has one row for each
event, and an arbitrary number of columns corresponding to different
features that were collected. Columns may be of type int, float, or str.
If an mne.Epochs object has a metadata attribute, you can select
subsets of epochs by using pandas query syntax directly. Here we’ll show
a few examples of how this looks.
# Authors: Chris Holdgraf <choldgraf@gmail.com>
# Jona Sassenhagen <jona.sassenhagen@gmail.com>
# Eric Larson <larson.eric.d@gmail.com>
# License: BSD (3-clause)
import os
import pandas as pd
import matplotlib.pyplot as plt
import mne
# First load some data
events = mne.read_events(os.path.join(mne.datasets.sample.data_path(),
'MEG/sample/sample_audvis_raw-eve.fif'))
raw = mne.io.read_raw_fif(os.path.join(mne.datasets.sample.data_path(),
'MEG/sample/sample_audvis_raw.fif'))
# We'll create some dummy names for each event type
event_id = {'Auditory/Left': 1, 'Auditory/Right': 2,
'Visual/Left': 3, 'Visual/Right': 4,
'smiley': 5, 'button': 32}
event_id_rev = {val: key for key, val in event_id.items()}
sides, kinds = [], []
for ev in events:
split = event_id_rev[ev[2]].lower().split('/')
if len(split) == 2:
kind, side = split
else:
kind = split[0]
side = 'both'
kinds.append(kind)
sides.append(side)
# Here's a helper function we'll use later
def plot_query_results(query):
fig = epochs[query].average().plot(show=False, time_unit='s')
title = fig.axes[0].get_title()
add = 'Query: {}\nNum Epochs: {}'.format(query, len(epochs[query]))
fig.axes[0].set(title='\n'.join([add, title]))
plt.show()
Out:
Opening raw data file /home/circleci/mne_data/MNE-sample-data/MEG/sample/sample_audvis_raw.fif...
Read a total of 3 projection items:
PCA-v1 (1 x 102) idle
PCA-v2 (1 x 102) idle
PCA-v3 (1 x 102) idle
Range : 25800 ... 192599 = 42.956 ... 320.670 secs
Ready.
First we’ll create our metadata object. This should be a
pandas.DataFrame with each row corresponding to an event.
Warning
Do not set or change the Dataframe index of epochs.metadata.
It will be controlled by MNE to mirror epochs.selection.
Also, while some inplace operations on epochs.metadata are
possible, do not manually drop or add rows, as this will
create inconsistency between the metadata and actual data.
metadata = {'event_time': events[:, 0] / raw.info['sfreq'],
'trial_number': range(len(events)),
'kind': kinds,
'side': sides}
metadata = pd.DataFrame(metadata)
metadata.head()
We can use this metadata object in the construction of an mne.Epochs
object. The metadata will then exist as an attribute:
epochs = mne.Epochs(raw, events, metadata=metadata, preload=True)
print(epochs.metadata.head())
Out:
Adding metadata with 4 columns
Replacing existing metadata with 4 columns
320 matching events found
Applying baseline correction (mode: mean)
Created an SSP operator (subspace dimension = 3)
3 projection items activated
Loading data for 320 events and 421 original time points ...
0 bad epochs dropped
event_time ... side
0 46.580589 ... right
1 47.193294 ... left
2 47.902567 ... left
3 48.648469 ... right
4 49.369397 ... right
[5 rows x 4 columns]
You can select rows by passing various queries to the Epochs object. For example, you can select a subset of events based on the value of a column.
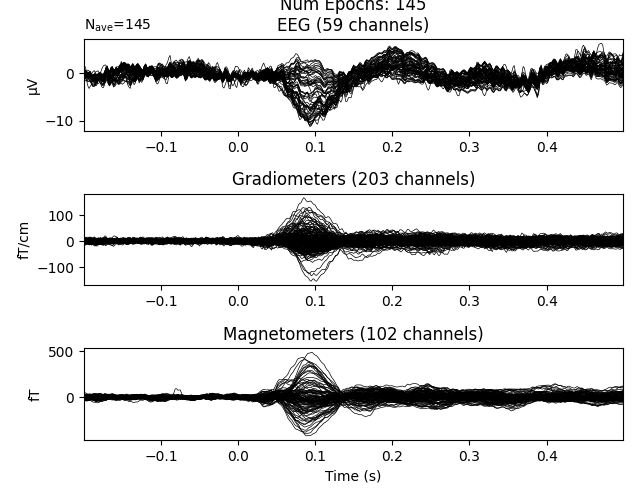
If a column has numeric values, you can also use numeric-style queries:
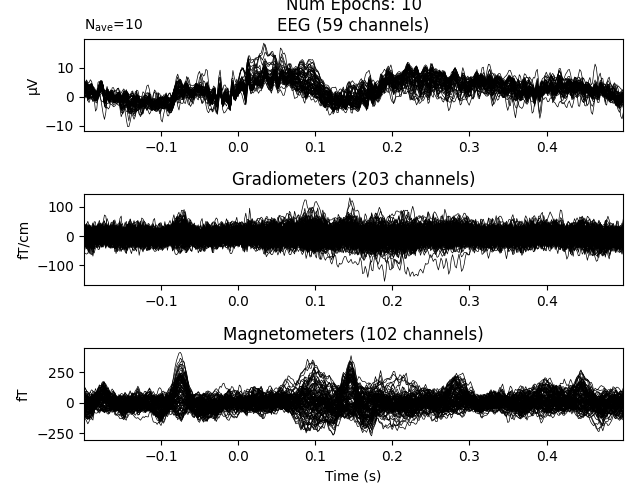
It is possible to chain these queries together, giving you more expressive ways to select particular epochs:
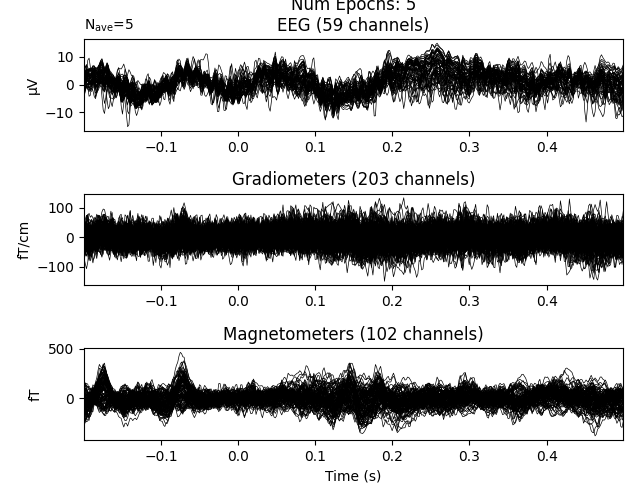
Any query that works with DataFrame.query will work for selecting epochs.
plot_events = ['smiley', 'button']
query = 'kind in {}'.format(plot_events)
plot_query_results(query)
![Query: kind in ['smiley', 'button'] Num Epochs: 31 EEG (59 channels), Gradiometers (203 channels), Magnetometers (102 channels)](../../_images/sphx_glr_plot_metadata_query_004.png)
Total running time of the script: ( 0 minutes 10.495 seconds)
Estimated memory usage: 570 MB