Note
Click here to download the full example code
Re-referencing the EEG signal¶
This example shows how to load raw data and apply some EEG referencing schemes.
# Authors: Marijn van Vliet <w.m.vanvliet@gmail.com>
# Alexandre Gramfort <alexandre.gramfort@inria.fr>
#
# License: BSD (3-clause)
import mne
from mne.datasets import sample
from matplotlib import pyplot as plt
print(__doc__)
# Setup for reading the raw data
data_path = sample.data_path()
raw_fname = data_path + '/MEG/sample/sample_audvis_filt-0-40_raw.fif'
event_fname = data_path + '/MEG/sample/sample_audvis_filt-0-40_raw-eve.fif'
event_id, tmin, tmax = 1, -0.2, 0.5
# Read the raw data
raw = mne.io.read_raw_fif(raw_fname, preload=True)
events = mne.read_events(event_fname)
# The EEG channels will be plotted to visualize the difference in referencing
# schemes.
picks = mne.pick_types(raw.info, meg=False, eeg=True, eog=True, exclude='bads')
Out:
Opening raw data file /home/circleci/mne_data/MNE-sample-data/MEG/sample/sample_audvis_filt-0-40_raw.fif...
Read a total of 4 projection items:
PCA-v1 (1 x 102) idle
PCA-v2 (1 x 102) idle
PCA-v3 (1 x 102) idle
Average EEG reference (1 x 60) idle
Range : 6450 ... 48149 = 42.956 ... 320.665 secs
Ready.
Reading 0 ... 41699 = 0.000 ... 277.709 secs...
We will now apply different EEG referencing schemes and plot the resulting
evoked potentials. Note that when we construct epochs with mne.Epochs, we
supply the proj=True argument. This means that any available projectors
are applied automatically. Specifically, if there is an average reference
projector set by raw.set_eeg_reference('average', projection=True), MNE
applies this projector when creating epochs.
reject = dict(eog=150e-6)
epochs_params = dict(events=events, event_id=event_id, tmin=tmin, tmax=tmax,
picks=picks, reject=reject, proj=True)
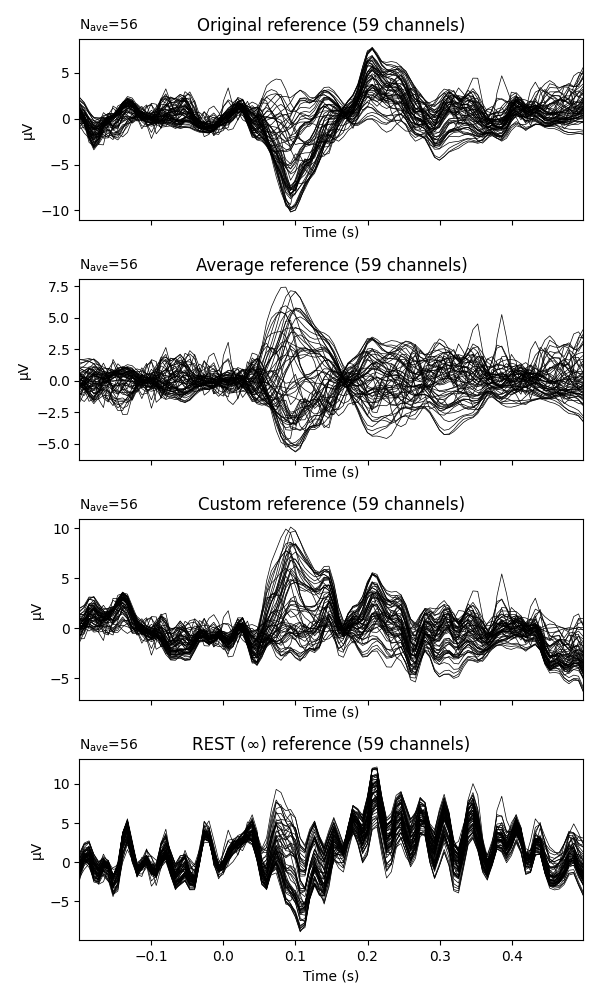
fig, (ax1, ax2, ax3, ax4) = plt.subplots(
nrows=4, ncols=1, sharex=True, figsize=(6, 10))
# We first want to plot the data without any added reference (i.e., using only
# the reference that was applied during recording of the data).
# However, this particular data already has an average reference projection
# applied that we now need to remove again using :func:`mne.set_eeg_reference`
raw, _ = mne.set_eeg_reference(raw, []) # use [] to remove average projection
evoked_no_ref = mne.Epochs(raw, **epochs_params).average()
evoked_no_ref.plot(axes=ax1, titles=dict(eeg='Original reference'), show=False,
time_unit='s')
# Now we want to plot the data with an average reference, so let's add the
# projection we removed earlier back to the data. Note that we can use
# "set_eeg_reference" as a method on the ``raw`` object as well.
raw.set_eeg_reference('average', projection=True)
evoked_car = mne.Epochs(raw, **epochs_params).average()
evoked_car.plot(axes=ax2, titles=dict(eeg='Average reference'), show=False,
time_unit='s')
# Re-reference from an average reference to the mean of channels EEG 001 and
# EEG 002.
raw.set_eeg_reference(['EEG 001', 'EEG 002'])
evoked_custom = mne.Epochs(raw, **epochs_params).average()
evoked_custom.plot(axes=ax3, titles=dict(eeg='Custom reference'),
time_unit='s', show=False)
# Re-reference using REST :footcite:`Yao2001`. To do this, we need a forward
# solution, which we can quickly create:
sphere = mne.make_sphere_model('auto', 'auto', raw.info)
src = mne.setup_volume_source_space(sphere=sphere, exclude=30.,
pos=15.) # large "pos" just for speed!
forward = mne.make_forward_solution(raw.info, trans=None, src=src, bem=sphere)
raw.set_eeg_reference('REST', forward=forward)
evoked_rest = mne.Epochs(raw, **epochs_params).average()
evoked_rest.plot(axes=ax4, titles=dict(eeg='REST (∞) reference'),
time_unit='s', show=True)
Out:
EEG channel type selected for re-referencing
EEG data marked as already having the desired reference.
Removing existing average EEG reference projection.
Created an SSP operator (subspace dimension = 3)
Not setting metadata
Not setting metadata
72 matching events found
Applying baseline correction (mode: mean)
3 projection items activated
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Removing projector <Projection | PCA-v1, active : True, n_channels : 102>
Removing projector <Projection | PCA-v2, active : True, n_channels : 102>
Removing projector <Projection | PCA-v3, active : True, n_channels : 102>
Adding average EEG reference projection.
1 projection items deactivated
Average reference projection was added, but has not been applied yet. Use the apply_proj method to apply it.
Not setting metadata
Not setting metadata
72 matching events found
Applying baseline correction (mode: mean)
Created an SSP operator (subspace dimension = 1)
4 projection items activated
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Removing projector <Projection | PCA-v1, active : True, n_channels : 102>
Removing projector <Projection | PCA-v2, active : True, n_channels : 102>
Removing projector <Projection | PCA-v3, active : True, n_channels : 102>
EEG channel type selected for re-referencing
Applying a custom EEG reference.
Removing existing average EEG reference projection.
Created an SSP operator (subspace dimension = 3)
Not setting metadata
Not setting metadata
72 matching events found
Applying baseline correction (mode: mean)
3 projection items activated
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Removing projector <Projection | PCA-v1, active : True, n_channels : 102>
Removing projector <Projection | PCA-v2, active : True, n_channels : 102>
Removing projector <Projection | PCA-v3, active : True, n_channels : 102>
Fitted sphere radius: 91.0 mm
Origin head coordinates: -4.1 16.0 51.7 mm
Origin device coordinates: 1.4 17.8 -10.3 mm
Equiv. model fitting -> RV = 0.00349028 %
mu1 = 0.944696 lambda1 = 0.137193
mu2 = 0.667458 lambda2 = 0.683737
mu3 = -0.26815 lambda3 = -0.0105603
Set up EEG sphere model with scalp radius 91.0 mm
Sphere : origin at (-4.1 16.0 51.7) mm
radius : 81.9 mm
grid : 15.0 mm
mindist : 5.0 mm
Exclude : 30.0 mm
Setting up the sphere...
Surface CM = ( -4.1 16.0 51.7) mm
Surface fits inside a sphere with radius 81.9 mm
Surface extent:
x = -86.0 ... 77.8 mm
y = -65.9 ... 97.9 mm
z = -30.2 ... 133.7 mm
Grid extent:
x = -90.0 ... 90.0 mm
y = -75.0 ... 105.0 mm
z = -45.0 ... 135.0 mm
2197 sources before omitting any.
655 sources after omitting infeasible sources not within 30.0 - 81.9 mm.
532 sources remaining after excluding the sources outside the surface and less than 5.0 mm inside.
Adjusting the neighborhood info.
Source space : MRI voxel -> MRI (surface RAS)
0.015000 0.000000 0.000000 -90.00 mm
0.000000 0.015000 0.000000 -75.00 mm
0.000000 0.000000 0.015000 -45.00 mm
0.000000 0.000000 0.000000 1.00
Source space : <SourceSpaces: [<discrete, n_used=532>] MRI (surface RAS) coords, ~575 kB>
MRI -> head transform : identity
Measurement data : instance of Info
Sphere model : origin at [-0.00413199 0.0159844 0.05174612] mm
Standard field computations
Do computations in head coordinates
Free source orientations
Read 1 source spaces a total of 532 active source locations
Coordinate transformation: MRI (surface RAS) -> head
1.000000 0.000000 0.000000 0.00 mm
0.000000 1.000000 0.000000 0.00 mm
0.000000 0.000000 1.000000 0.00 mm
0.000000 0.000000 0.000000 1.00
Read 306 MEG channels from info
99 coil definitions read
Coordinate transformation: MEG device -> head
0.991420 -0.039936 -0.124467 -6.13 mm
0.060661 0.984012 0.167456 0.06 mm
0.115790 -0.173570 0.977991 64.74 mm
0.000000 0.000000 0.000000 1.00
MEG coil definitions created in head coordinates.
Read 60 EEG channels from info
Head coordinate coil definitions created.
Source spaces are now in head coordinates.
Using the sphere model.
Setting up compensation data...
No compensation set. Nothing more to do.
Using the equivalent source approach in the homogeneous sphere for EEG
Computing MEG at 532 source locations (free orientations)...
Computing EEG at 532 source locations (free orientations)...
Finished.
EEG channel type selected for re-referencing
Applying REST reference.
Applying a custom EEG reference.
Created an SSP operator (subspace dimension = 3)
59 out of 366 channels remain after picking
Not setting metadata
Not setting metadata
72 matching events found
Applying baseline correction (mode: mean)
3 projection items activated
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Removing projector <Projection | PCA-v1, active : True, n_channels : 102>
Removing projector <Projection | PCA-v2, active : True, n_channels : 102>
Removing projector <Projection | PCA-v3, active : True, n_channels : 102>
References¶
Total running time of the script: ( 0 minutes 3.316 seconds)
Estimated memory usage: 237 MB