Note
Click here to download the full example code
Creating MNE’s data structures from scratch¶
MNE provides mechanisms for creating various core objects directly from NumPy arrays.
import mne
import numpy as np
Creating Info objects¶
Note
for full documentation on the Info object, see
The Info data structure. See also Creating MNE objects from data arrays.
Normally, mne.Info objects are created by the various
data import functions.
However, if you wish to create one from scratch, you can use the
mne.create_info() function to initialize the minimally required
fields. Further fields can be assigned later as one would with a regular
dictionary.
The following creates the absolute minimum info structure:
# Create some dummy metadata
n_channels = 32
sampling_rate = 200
info = mne.create_info(n_channels, sampling_rate)
print(info)
Out:
<Info | 7 non-empty values
bads: []
ch_names: 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, ...
chs: 32 MISC
custom_ref_applied: False
highpass: 0.0 Hz
lowpass: 100.0 Hz
meas_date: unspecified
nchan: 32
projs: []
sfreq: 200.0 Hz
>
You can also supply more extensive metadata:
# Names for each channel
channel_names = ['MEG1', 'MEG2', 'Cz', 'Pz', 'EOG']
# The type (mag, grad, eeg, eog, misc, ...) of each channel
channel_types = ['grad', 'grad', 'eeg', 'eeg', 'eog']
# The sampling rate of the recording
sfreq = 1000 # in Hertz
# The EEG channels use the standard naming strategy.
# By supplying the 'montage' parameter, approximate locations
# will be added for them
montage = 'standard_1005'
# Initialize required fields
info = mne.create_info(channel_names, sfreq, channel_types)
info.set_montage(montage)
# Add some more information
info['description'] = 'My custom dataset'
info['bads'] = ['Pz'] # Names of bad channels
print(info)
Out:
<Info | 10 non-empty values
bads: 1 items (Pz)
ch_names: MEG1, MEG2, Cz, Pz, EOG
chs: 2 GRAD, 2 EEG, 1 EOG
custom_ref_applied: False
description: My custom dataset
dig: 5 items (3 Cardinal, 2 EEG)
highpass: 0.0 Hz
lowpass: 500.0 Hz
meas_date: unspecified
nchan: 5
projs: []
sfreq: 1000.0 Hz
>
Note
When assigning new values to the fields of an
mne.Info object, it is important that the
fields are consistent:
The length of the channel information field
chsmust benchan.The length of the
ch_namesfield must benchan.The
ch_namesfield should be consistent with thenamefield of the channel information contained inchs.
Creating Raw objects¶
To create a mne.io.Raw object from scratch, you can use the
mne.io.RawArray class, which implements raw data that is backed by a
numpy array. The correct units for the data are:
V: eeg, eog, seeg, emg, ecg, bio, ecog
T: mag
T/m: grad
M: hbo, hbr
Am: dipole
AU: misc
The mne.io.RawArray constructor simply takes the data matrix and
mne.Info object:
# Generate some random data
data = np.random.randn(5, 1000)
# Initialize an info structure
info = mne.create_info(
ch_names=['MEG1', 'MEG2', 'EEG1', 'EEG2', 'EOG'],
ch_types=['grad', 'grad', 'eeg', 'eeg', 'eog'],
sfreq=100)
custom_raw = mne.io.RawArray(data, info)
print(custom_raw)
Out:
Creating RawArray with float64 data, n_channels=5, n_times=1000
Range : 0 ... 999 = 0.000 ... 9.990 secs
Ready.
<RawArray | 5 x 1000 (10.0 s), ~53 kB, data loaded>
Creating Epochs objects¶
To create an mne.Epochs object from scratch, you can use the
mne.EpochsArray class, which uses a numpy array directly without
wrapping a raw object. The array must be of shape
(n_epochs, n_chans, n_times). The proper units of measure are listed
above.
# Generate some random data: 10 epochs, 5 channels, 2 seconds per epoch
sfreq = 100
data = np.random.randn(10, 5, sfreq * 2)
# Initialize an info structure
info = mne.create_info(
ch_names=['MEG1', 'MEG2', 'EEG1', 'EEG2', 'EOG'],
ch_types=['grad', 'grad', 'eeg', 'eeg', 'eog'],
sfreq=sfreq)
It is necessary to supply an “events” array in order to create an Epochs
object. This is of shape (n_events, 3) where the first column is the
sample number (time) of the event, the second column indicates the value from
which the transition is made from (only used when the new value is bigger
than the old one), and the third column is the new event value.
More information about the event codes: subject was either smiling or frowning
event_id = dict(smiling=1, frowning=2)
Finally, we must specify the beginning of an epoch (the end will be inferred from the sampling frequency and n_samples)
# Trials were cut from -0.1 to 1.0 seconds
tmin = -0.1
Now we can create the mne.EpochsArray object
custom_epochs = mne.EpochsArray(data, info, events, tmin, event_id)
print(custom_epochs)
# We can treat the epochs object as we would any other
_ = custom_epochs['smiling'].average().plot(time_unit='s')
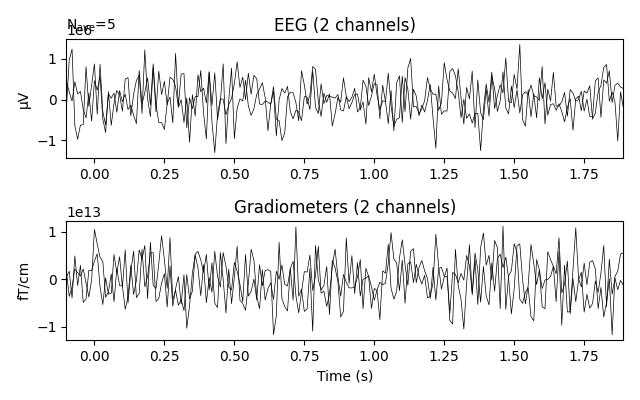
Out:
Not setting metadata
Not setting metadata
10 matching events found
No baseline correction applied
0 projection items activated
0 bad epochs dropped
<EpochsArray | 10 events (all good), -0.1 - 1.89 sec, baseline off, ~92 kB, data loaded,
'frowning': 5
'smiling': 5>
Need more than one channel to make topography for grad. Disabling interactivity.
Creating Evoked Objects¶
If you already have data that is collapsed across trials, you may also
directly create an evoked array. Its constructor accepts an array of shape
(n_chans, n_times) in addition to some bookkeeping parameters.
The proper units of measure for the data are listed above.
# The averaged data
data_evoked = data.mean(0)
# The number of epochs that were averaged
nave = data.shape[0]
# A comment to describe to evoked (usually the condition name)
comment = "Smiley faces"
# Create the Evoked object
evoked_array = mne.EvokedArray(data_evoked, info, tmin,
comment=comment, nave=nave)
print(evoked_array)
_ = evoked_array.plot(time_unit='s')
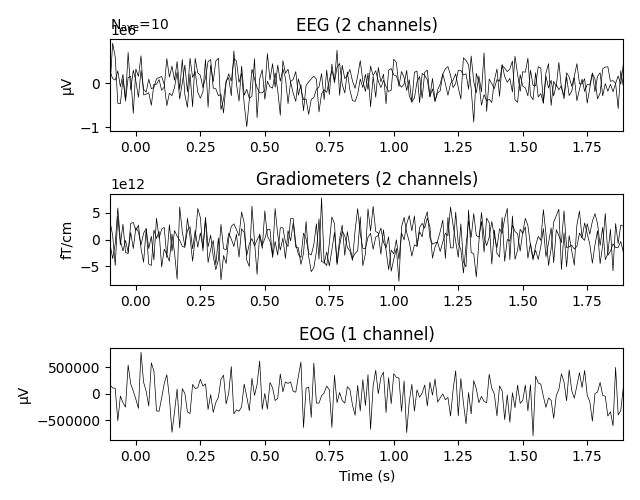
Out:
<Evoked | 'Smiley faces' (average, N=10), [-0.1, 1.89] sec, 5 ch, ~22 kB>
Need more than one channel to make topography for grad. Disabling interactivity.
Total running time of the script: ( 0 minutes 8.040 seconds)
Estimated memory usage: 8 MB