Note
Click here to download the full example code or to run this example in your browser via Binder
08. Convert iEEG data to BIDS format¶
In this example, we use MNE-BIDS to create a BIDS-compatible directory of iEEG data. Specifically, we will follow these steps:
Download some iEEG data from the PhysioBank database and the MNE-ECoG ex.
Load the data, extract information, and save in a new BIDS directory.
Check the result and compare it with the standard.
Confirm that written iEEG coordinates are the same before
write_raw_bids()was called.
The iEEG data will be written by write_raw_bids() with
the addition of extra metadata elements in the following files:
sidecar.json
electrodes.tsv
coord_system.json
events.tsv
channels.tsv
Compared to EEG data, the main differences are within the coord_system and electrodes files. For more information on these files, refer to the iEEG-BIDS specification.
Step 1: Download the data¶
First, we need some data to work with. We will use the data downloaded via MNE-Python’s API.
https://mne.tools/stable/generated/mne.datasets.misc.data_path.
Conveniently, there is already a data loading function available with MNE-Python:
# get MNE directory w/ example data
mne_dir = mne.get_config('MNE_DATASETS_SAMPLE_PATH')
# The electrode coords data are in the Matlab format: '.mat'.
# This is easy to read in with :func:`scipy.io.loadmat` function.
mat = loadmat(mne.datasets.misc.data_path() + '/ecog/sample_ecog.mat')
ch_names = mat['ch_names'].tolist()
ch_names = [x.strip() for x in ch_names]
elec = mat['elec'] # electrode positions given in meters
Now we make a montage stating that the iEEG contacts are in MRI coordinate system.
Out:
Created 64 channel positions
{'POL LPG1-Ref': array([-0.01319475, -0.06537354, 0.07015674]), 'POL LPG2-Ref': array([-0.01446107, -0.05939256, 0.07805224]), 'POL LPG3-Ref': array([-0.01562162, -0.05178238, 0.08383159]), 'POL LPG4-Ref': array([-0.01542273, -0.04525053, 0.08948554]), 'POL LPG5-Ref': array([-0.01487775, -0.03852912, 0.09396847]), 'POL LPG6-Ref': array([-0.01307837, -0.02902713, 0.09850599]), 'POL LPG7-Ref': array([-0.01359957, -0.02028753, 0.1009258 ]), 'POL LPG8-Ref': array([-0.0108829 , -0.00930631, 0.10348425]), 'POL LPG9-Ref': array([-0.02154753, -0.06642563, 0.06912147]), 'POL LPG10-Ref': array([-0.02327222, -0.06074122, 0.07691736]), 'POL LPG11-Ref': array([-0.02474644, -0.0533903 , 0.08223433]), 'POL LPG12-Ref': array([-0.02632337, -0.04606533, 0.08709487]), 'POL LPG13-Ref': array([-0.0261479 , -0.0382572 , 0.09175405]), 'POL LPG14-Ref': array([-0.02525023, -0.02987171, 0.09489567]), 'POL LPG15-Ref': array([-0.02331042, -0.01934099, 0.09894071]), 'POL LPG16-Ref': array([-0.02108674, -0.00797552, 0.1025569 ]), 'POL LPG17-Ref': array([-0.0303566 , -0.06742905, 0.06775238]), 'POL LPG18-Ref': array([-0.03277563, -0.06100421, 0.0737788 ]), 'POL LPG19-Ref': array([-0.03369498, -0.05385209, 0.07930177]), 'POL LPG20-Ref': array([-0.03437536, -0.04655476, 0.08423193]), 'POL LPG21-Ref': array([-0.03627066, -0.03878722, 0.08779606]), 'POL LPG22-Ref': array([-0.03445379, -0.02883017, 0.09262682]), 'POL LPG23-Ref': array([-0.03352449, -0.01795435, 0.0954834 ]), 'POL LPG24-Ref': array([-0.02986222, -0.00800269, 0.09773305]), 'POL LPG25-Ref': array([-0.03949066, -0.06611425, 0.06322438]), 'POL LPG26-Ref': array([-0.04173993, -0.05987147, 0.06903918]), 'POL LPG27-Ref': array([-0.04263702, -0.05290591, 0.0743644 ]), 'POL LPG28-Ref': array([-0.04377296, -0.04564748, 0.07952718]), 'POL LPG29-Ref': array([-0.04569133, -0.03760912, 0.08295954]), 'POL LPG30-Ref': array([-0.04460317, -0.02733297, 0.08814251]), 'POL LPG31-Ref': array([-0.04300759, -0.01760467, 0.09012438]), 'POL LPG32-Ref': array([-0.03956011, -0.00625048, 0.08837144]), 'POL LPG33-Ref': array([-0.0445793 , -0.06573858, 0.05814573]), 'POL LPG34-Ref': array([-0.04824952, -0.05929582, 0.06322013]), 'POL LPG35-Ref': array([-0.05054796, -0.05233206, 0.06939953]), 'POL LPG36-Ref': array([-0.05363233, -0.04382743, 0.07313115]), 'POL LPG37-Ref': array([-0.05484415, -0.03568853, 0.07694293]), 'POL LPG38-Ref': array([-0.05327717, -0.02539124, 0.08083972]), 'POL LPG39-Ref': array([-0.05115428, -0.01564116, 0.08229832]), 'POL LPG40-Ref': array([-0.04732043, -0.00440023, 0.08078417]), 'POL LPG41-Ref': array([-0.05180684, -0.06374008, 0.05126565]), 'POL LPG42-Ref': array([-0.054985 , -0.05721633, 0.05740975]), 'POL LPG43-Ref': array([-0.05751256, -0.05005348, 0.06253482]), 'POL LPG44-Ref': array([-0.06058705, -0.0414252 , 0.06662151]), 'POL LPG45-Ref': array([-0.06140005, -0.03339784, 0.07086563]), 'POL LPG46-Ref': array([-0.06117977, -0.02303283, 0.07327374]), 'POL LPG47-Ref': array([-0.05892394, -0.01350239, 0.074307 ]), 'POL LPG48-Ref': array([-0.05525155, -0.00254127, 0.0739286 ]), 'POL LPG49-Ref': array([-0.05753089, -0.0605335 , 0.04550514]), 'POL LPG50-Ref': array([-0.06154114, -0.05356016, 0.0499875 ]), 'POL LPG51-Ref': array([-0.06351517, -0.0473602 , 0.05540717]), 'POL LPG52-Ref': array([-0.06602076, -0.03851005, 0.0584159 ]), 'POL LPG53-Ref': array([-0.06714931, -0.0305876 , 0.06297414]), 'POL LPG54-Ref': array([-0.06722665, -0.02039802, 0.06437857]), 'POL LPG55-Ref': array([-0.06426395, -0.01123459, 0.06384582]), 'POL LPG56-Ref': array([-0.06023369, -0.00067103, 0.0640136 ]), 'POL LPG57-Ref': array([-0.06137646, -0.05744936, 0.03716252]), 'POL LPG58-Ref': array([-0.06594827, -0.04996002, 0.04119299]), 'POL LPG59-Ref': array([-0.06806244, -0.04335446, 0.04628402]), 'POL LPG60-Ref': array([-0.0694307 , -0.0357257 , 0.05029811]), 'POL LPG61-Ref': array([-0.0704345 , -0.02766433, 0.05298704]), 'POL LPG62-Ref': array([-0.07051575, -0.01773528, 0.0540562 ]), 'POL LPG63-Ref': array([-0.06801403, -0.00742642, 0.05301937]), 'POL LPG64-Ref': array([-0.06415953, 0.00155849, 0.05387343])}
We also need to get some sample iEEG data, so we will just generate random data from white noise. Here is where you would use your own data if you had it.
ieegdata = np.random.rand(len(ch_names), 1000)
# We will create a :func:`mne.io.RawArray` object and
# use the montage we created.
info = mne.create_info(ch_names, 1000., 'ecog')
raw = mne.io.RawArray(ieegdata, info)
raw.set_montage(montage)
Out:
Creating RawArray with float64 data, n_channels=64, n_times=1000
Range : 0 ... 999 = 0.000 ... 0.999 secs
Ready.
/home/stefanappelhoff/Desktop/bids/mne-bids/examples/convert_ieeg_to_bids.py:90: RuntimeWarning: Fiducial point nasion not found, assuming identity unknown to head transformation
raw.set_montage(montage)
<RawArray | 64 x 1000 (1.0 s), ~676 kB, data loaded>
Let us confirm what our channel coordinates look like.
# make a plot of the sensors in 2D plane
raw.plot_sensors(ch_type='ecog')
# Get the first 5 channels and show their locations.
picks = mne.pick_types(raw.info, ecog=True)
dig = [raw.info['dig'][pick] for pick in picks]
chs = [raw.info['chs'][pick] for pick in picks]
pos = np.array([ch['r'] for ch in dig[:5]])
ch_names = np.array([ch['ch_name'] for ch in chs[:5]])
print("The channel coordinates before writing into BIDS: ")
pprint([x for x in zip(ch_names, pos)])
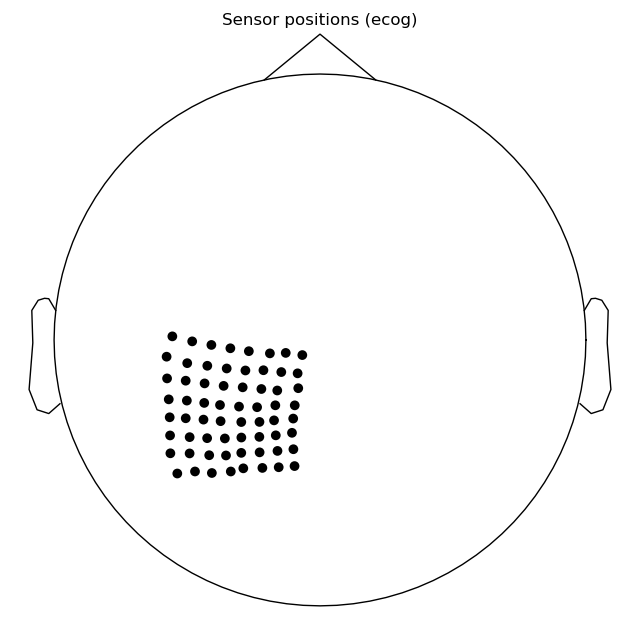
Out:
The channel coordinates before writing into BIDS:
[('POL LPG1-Ref', array([-0.01319475, -0.06537354, 0.07015674])),
('POL LPG2-Ref', array([-0.02327222, -0.06074122, 0.07691736])),
('POL LPG3-Ref', array([-0.02474644, -0.0533903 , 0.08223433])),
('POL LPG4-Ref', array([-0.02632337, -0.04606533, 0.08709487])),
('POL LPG5-Ref', array([-0.0261479 , -0.0382572 , 0.09175405]))]
Step 2: Formatting as BIDS¶
Now, let us format the Raw object into BIDS.
With this step, we have everything to start a new BIDS directory using
our data. To do that, we can use write_raw_bids()
Generally, write_raw_bids() tries to extract as much
meta data as possible from the raw data and then formats it in a BIDS
compatible way. write_raw_bids() takes a bunch of inputs, most of
which are however optional. The required inputs are:
rawbids_basenamebids_root
… as you can see in the docstring:
print(write_raw_bids.__doc__)
Out:
Save raw data to a BIDS-compliant folder structure.
.. warning:: * The original file is simply copied over if the original
file format is BIDS-supported for that modality. Otherwise,
this function will convert to a BIDS-supported file format
while warning the user. For EEG and iEEG data, conversion
will be to BrainVision format, for MEG conversion will be
to FIF.
* ``mne-bids`` will infer the manufacturer information
from the file extension. If your file format is non-standard
for the manufacturer, please update the manufacturer field
in the sidecars manually.
Parameters
----------
raw : instance of mne.io.Raw
The raw data. It must be an instance of mne.Raw. The data should not be
loaded from disk, i.e., raw.preload must be False.
bids_basename : str
The base filename of the BIDS compatible files. Typically, this can be
generated using make_bids_basename.
Example: `sub-01_ses-01_task-testing_acq-01_run-01`.
This will write the following files in the correct subfolder of the
bids_root::
sub-01_ses-01_task-testing_acq-01_run-01_meg.fif
sub-01_ses-01_task-testing_acq-01_run-01_meg.json
sub-01_ses-01_task-testing_acq-01_run-01_channels.tsv
sub-01_ses-01_task-testing_acq-01_run-01_coordsystem.json
and the following one if events_data is not None::
sub-01_ses-01_task-testing_acq-01_run-01_events.tsv
and add a line to the following files::
participants.tsv
scans.tsv
Note that the modality 'meg' is automatically inferred from the raw
object and extension '.fif' is copied from raw.filenames.
bids_root : str | pathlib.Path
The path of the root of the BIDS compatible folder. The session and
subject specific folders will be populated automatically by parsing
bids_basename.
events_data : str | pathlib.Path | array | None
The events file. If a string or a Path object, specifies the path of
the events file. If an array, the MNE events array (shape n_events, 3).
If None, events will be inferred from the stim channel using
`mne.find_events`.
event_id : dict | None
The event id dict used to create a 'trial_type' column in events.tsv
anonymize : dict | None
If None is provided (default) no anonymization is performed.
If a dictionary is passed, data will be anonymized; identifying data
structures such as study date and time will be changed.
`daysback` is a required argument and `keep_his` is optional, these
arguments are passed to :func:`mne.io.anonymize_info`.
`daysback` : int
Number of days to move back the date. To keep relative dates for
a subject use the same `daysback`. According to BIDS
specifications, the number of days back must be great enough
that the date is before 1925.
`keep_his` : bool
If True info['subject_info']['his_id'] of subject_info will NOT be
overwritten. Defaults to False.
overwrite : bool
Whether to overwrite existing files or data in files.
Defaults to False.
If overwrite is True, any existing files with the same BIDS parameters
will be overwritten with the exception of the `participants.tsv` and
`scans.tsv` files. For these files, parts of pre-existing data that
match the current data will be replaced.
If overwrite is False, no existing data will be overwritten or
replaced.
verbose : bool
If verbose is True, this will print a snippet of the sidecar files. If
False, no content will be printed.
Returns
-------
bids_root : str
The path of the root of the BIDS compatible folder.
Notes
-----
For the participants.tsv file, the raw.info['subject_info'] should be
updated and raw.info['meas_date'] should not be None to compute the age
of the participant correctly.
Let us initialize some of the necessary data for the subject There is a subject, and specific task for the dataset.
subject_id = '001' # zero padding to account for >100 subjects in this dataset
task = 'testresteyes'
# There is the root directory for where we will write our data.
bids_root = os.path.join(mne_dir, 'ieegmmidb_bids')
Now we just need to specify a few iEEG details to make things work: We need the basename of the dataset. In addition, write_raw_bids requires a filenames of the Raw object to be non-empty, so since we initialized the dataset from an array, we need to do a hack where we temporarily save the data to disc before reading it back in.
# Now convert our data to be in a new BIDS dataset.
bids_basename = make_bids_basename(subject=subject_id,
task=task,
acquisition="ecog")
# need to set the filenames if we are initializing data from array
with tempfile.TemporaryDirectory() as tmp_root:
tmp_fpath = os.path.join(tmp_root, "test_raw.fif")
raw.save(tmp_fpath)
raw = mne.io.read_raw_fif(tmp_fpath)
# write `raw` to BIDS and anonymize it into BrainVision format
write_raw_bids(raw, bids_basename, bids_root=bids_root,
anonymize=dict(daysback=30000), overwrite=True)
Out:
Writing /tmp/tmp0q32kptc/test_raw.fif
Closing /tmp/tmp0q32kptc/test_raw.fif [done]
Opening raw data file /tmp/tmp0q32kptc/test_raw.fif...
Range : 0 ... 999 = 0.000 ... 0.999 secs
Ready.
Opening raw data file /tmp/tmp0q32kptc/test_raw.fif...
Range : 0 ... 999 = 0.000 ... 0.999 secs
Ready.
Creating folder: /home/stefanappelhoff/mne_data/ieegmmidb_bids/sub-001/ieeg
/home/stefanappelhoff/Desktop/bids/mne-bids/mne_bids/write.py:1231: RuntimeWarning: Input info has 'meas_date' set to None. Removing all information from time/date structures. *NOT* performing any time shifts
keep_his=keep_his)
/home/stefanappelhoff/Desktop/bids/mne-bids/mne_bids/write.py:1238: UserWarning: Converting to BV for anonymization
warn('Converting to BV for anonymization')
Writing '/home/stefanappelhoff/mne_data/ieegmmidb_bids/participants.tsv'...
participant_id age sex hand
sub-001 n/a n/a n/a
Writing '/home/stefanappelhoff/mne_data/ieegmmidb_bids/participants.json'...
{
"participant_id": {
"Description": "Unique participant identifier"
},
"age": {
"Description": "Age of the participant at time of testing",
"Units": "years"
},
"sex": {
"Description": "Biological sex of the participant",
"Levels": {
"F": "female",
"M": "male"
}
},
"hand": {
"Description": "Handedness of the participant",
"Levels": {
"R": "right",
"L": "left",
"A": "ambidextrous"
}
}
}
Writing '/home/stefanappelhoff/mne_data/ieegmmidb_bids/sub-001/sub-001_scans.tsv'...
filename acq_time
ieeg/sub-001_task-testresteyes_acq-ecog_ieeg.vhdr n/a
Writing '/home/stefanappelhoff/mne_data/ieegmmidb_bids/sub-001/ieeg/sub-001_acq-ecog_electrodes.tsv'...
name x y z size
POL LPG1-Ref -0.013194745406508446 -0.06537354737520218 0.07015674561262131 n/a
POL LPG2-Ref -0.014461069367825985 -0.05939256399869919 0.07805224508047104 n/a
POL LPG3-Ref -0.015621616505086422 -0.05178237706422806 0.08383159339427948 n/a
POL LPG4-Ref -0.015422730706632137 -0.045250535011291504 0.0894855409860611 n/a
POL LPG5-Ref -0.014877748675644398 -0.03852912411093712 0.093968465924263 n/a
Writing '/home/stefanappelhoff/mne_data/ieegmmidb_bids/sub-001/ieeg/sub-001_acq-ecog_coordsystem.json'...
{
"iEEGCoordinateSystem": "mri",
"iEEGCoordinateUnits": "m"
}
/home/stefanappelhoff/Desktop/bids/mne-bids/mne_bids/utils.py:460: UserWarning: No events found or provided. Please make sure to set channel type using raw.set_channel_types or provide events_data.
warnings.warn('No events found or provided. Please make sure to'
Writing '/home/stefanappelhoff/mne_data/ieegmmidb_bids/dataset_description.json'...
{
"Name": " ",
"BIDSVersion": "1.2.2",
"Authors": [
"MNE-BIDS"
]
}
Effective window size : 0.256 (s)
/home/stefanappelhoff/Desktop/bids/mne-bids/mne_bids/write.py:559: UserWarning: No line frequency found, defaulting to 60 Hz estimated from multi-taper FFT on 10 seconds of data.
'on 10 seconds of data.'.format(powerlinefrequency))
Writing '/home/stefanappelhoff/mne_data/ieegmmidb_bids/sub-001/ieeg/sub-001_task-testresteyes_acq-ecog_ieeg.json'...
{
"TaskName": "testresteyes",
"Manufacturer": "Elekta",
"PowerLineFrequency": 60,
"SamplingFrequency": 1000.0,
"SoftwareFilters": "n/a",
"RecordingDuration": 0.999,
"RecordingType": "continuous",
"iEEGReference": "n/a",
"ECOGChannelCount": 64,
"SEEGChannelCount": 0,
"EEGChannelCount": 0,
"EOGChannelCount": 0,
"ECGChannelCount": 0,
"EMGChannelCount": 0,
"MiscChannelCount": 0,
"TriggerChannelCount": 0
}
Writing '/home/stefanappelhoff/mne_data/ieegmmidb_bids/sub-001/ieeg/sub-001_task-testresteyes_acq-ecog_channels.tsv'...
name type units low_cutoff high_cutoff description sampling_frequency status
POL LPG1-Ref ECOG V 0.0 500.0 Electrocorticography 1000.0 good
POL LPG2-Ref ECOG V 0.0 500.0 Electrocorticography 1000.0 good
POL LPG3-Ref ECOG V 0.0 500.0 Electrocorticography 1000.0 good
POL LPG4-Ref ECOG V 0.0 500.0 Electrocorticography 1000.0 good
POL LPG5-Ref ECOG V 0.0 500.0 Electrocorticography 1000.0 good
/home/stefanappelhoff/Desktop/bids/mne-bids/mne_bids/write.py:1324: UserWarning: Converting data files to BrainVision format
warn('Converting data files to BrainVision format')
Step 3: Check and compare with standard¶
# Now we have written our BIDS directory.
print_dir_tree(bids_root)
Out:
|ieegmmidb_bids/
|--- participants.tsv
|--- dataset_description.json
|--- participants.json
|--- sub-001/
|------ sub-001_scans.tsv
|------ ieeg/
|--------- sub-001_task-testresteyes_acq-ecog_ieeg.vmrk
|--------- sub-001_task-testresteyes_acq-ecog_ieeg.vhdr
|--------- sub-001_task-testresteyes_acq-ecog_channels.tsv
|--------- sub-001_acq-ecog_electrodes.tsv
|--------- sub-001_task-testresteyes_acq-ecog_ieeg.json
|--------- sub-001_task-testresteyes_acq-ecog_ieeg.eeg
|--------- sub-001_acq-ecog_coordsystem.json
MNE-BIDS has created a suitable directory structure for us, and among other meta data files, it started an events.tsv and channels.tsv and made an initial dataset_description.json on top!
Now it’s time to manually check the BIDS directory and the meta files to add all the information that MNE-BIDS could not infer. For instance, you must describe iEEGReference and iEEGGround yourself. It’s easy to find these by searching for “n/a” in the sidecar files.
$ grep -i ‘n/a’ <bids_root>
Remember that there is a convenient javascript tool to validate all your BIDS directories called the “BIDS-validator”, available as a web version and a command line tool:
Web version: https://bids-standard.github.io/bids-validator/
Command line tool: https://www.npmjs.com/package/bids-validator
Step 4: Plot output channels and check that they match!¶
Now we have written our BIDS directory. We can use
read_raw_bids() to read in the data.
# read in the BIDS dataset and plot the coordinates
bids_fname = bids_basename + "_ieeg.vhdr"
raw = read_raw_bids(bids_fname, bids_root=bids_root)
# get the first 5 channels and show their locations
# this should match what was printed earlier.
picks = mne.pick_types(raw.info, ecog=True)
dig = [raw.info['dig'][pick] for pick in picks]
chs = [raw.info['chs'][pick] for pick in picks]
pos = np.array([ch['r'] for ch in dig[:5]])
ch_names = np.array([ch['ch_name'] for ch in chs[:5]])
print("The channel coordinates after writing into BIDS: ")
pprint([x for x in zip(ch_names, pos)])
# make a plot of the sensors in 2D plane
raw.plot_sensors(ch_type='ecog')
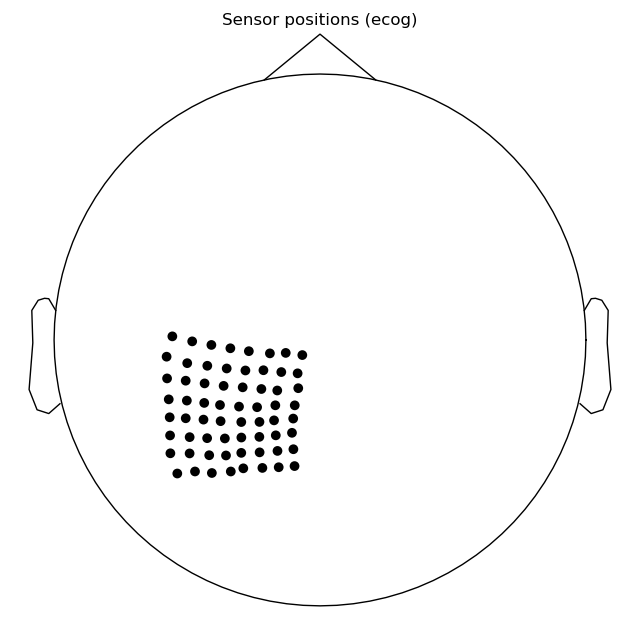
Out:
Extracting parameters from /home/stefanappelhoff/mne_data/ieegmmidb_bids/sub-001/ieeg/sub-001_task-testresteyes_acq-ecog_ieeg.vhdr...
Setting channel info structure...
/home/stefanappelhoff/Desktop/bids/mne-bids/mne_bids/utils.py:635: UserWarning: Did not find any events.tsv file associated with sub-001_task-testresteyes_acq-ecog_ieeg.vhdr.
The search_str was "/home/stefanappelhoff/mne_data/ieegmmidb_bids/sub-001/**/sub-001*events.tsv"
warnings.warn(msg)
Reading channel info from /home/stefanappelhoff/mne_data/ieegmmidb_bids/sub-001/ieeg/sub-001_task-testresteyes_acq-ecog_channels.tsv.
Reading electrode coords from /home/stefanappelhoff/mne_data/ieegmmidb_bids/sub-001/ieeg/sub-001_acq-ecog_electrodes.tsv.
The read in electrodes file is:
OrderedDict([('name', ['POL LPG1-Ref', 'POL LPG2-Ref', 'POL LPG3-Ref', 'POL LPG4-Ref', 'POL LPG5-Ref', 'POL LPG6-Ref', 'POL LPG7-Ref', 'POL LPG8-Ref', 'POL LPG9-Ref', 'POL LPG10-Ref', 'POL LPG11-Ref', 'POL LPG12-Ref', 'POL LPG13-Ref', 'POL LPG14-Ref', 'POL LPG15-Ref', 'POL LPG16-Ref', 'POL LPG17-Ref', 'POL LPG18-Ref', 'POL LPG19-Ref', 'POL LPG20-Ref', 'POL LPG21-Ref', 'POL LPG22-Ref', 'POL LPG23-Ref', 'POL LPG24-Ref', 'POL LPG25-Ref', 'POL LPG26-Ref', 'POL LPG27-Ref', 'POL LPG28-Ref', 'POL LPG29-Ref', 'POL LPG30-Ref', 'POL LPG31-Ref', 'POL LPG32-Ref', 'POL LPG33-Ref', 'POL LPG34-Ref', 'POL LPG35-Ref', 'POL LPG36-Ref', 'POL LPG37-Ref', 'POL LPG38-Ref', 'POL LPG39-Ref', 'POL LPG40-Ref', 'POL LPG41-Ref', 'POL LPG42-Ref', 'POL LPG43-Ref', 'POL LPG44-Ref', 'POL LPG45-Ref', 'POL LPG46-Ref', 'POL LPG47-Ref', 'POL LPG48-Ref', 'POL LPG49-Ref', 'POL LPG50-Ref', 'POL LPG51-Ref', 'POL LPG52-Ref', 'POL LPG53-Ref', 'POL LPG54-Ref', 'POL LPG55-Ref', 'POL LPG56-Ref', 'POL LPG57-Ref', 'POL LPG58-Ref', 'POL LPG59-Ref', 'POL LPG60-Ref', 'POL LPG61-Ref', 'POL LPG62-Ref', 'POL LPG63-Ref', 'POL LPG64-Ref']), ('x', ['-0.013194745406508446', '-0.014461069367825985', '-0.015621616505086422', '-0.015422730706632137', '-0.014877748675644398', '-0.013078372925519943', '-0.013599568046629429', '-0.010882899165153503', '-0.021547531709074974', '-0.023272216320037842', '-0.02474643662571907', '-0.02632337249815464', '-0.02614789642393589', '-0.025250233709812164', '-0.02331041917204857', '-0.021086739376187325', '-0.030356602743268013', '-0.032775625586509705', '-0.03369498252868652', '-0.03437535837292671', '-0.036270663142204285', '-0.03445379436016083', '-0.03352448716759682', '-0.02986222319304943', '-0.03949065878987312', '-0.04173993319272995', '-0.042637016624212265', '-0.04377295821905136', '-0.04569132626056671', '-0.04460316523909569', '-0.043007589876651764', '-0.03956010937690735', '-0.04457930102944374', '-0.048249516636133194', '-0.05054796114563942', '-0.053632333874702454', '-0.054844148457050323', '-0.05327717587351799', '-0.05115427449345589', '-0.047320425510406494', '-0.05180684104561806', '-0.05498500168323517', '-0.0575125627219677', '-0.060587044805288315', '-0.061400044709444046', '-0.06117977201938629', '-0.058923933655023575', '-0.055251553654670715', '-0.05753088742494583', '-0.06154113635420799', '-0.06351517140865326', '-0.06602075695991516', '-0.06714931130409241', '-0.06722664833068848', '-0.06426394730806351', '-0.06023368984460831', '-0.06137646362185478', '-0.06594827026128769', '-0.0680624470114708', '-0.06943069398403168', '-0.07043450325727463', '-0.07051574438810349', '-0.06801402568817139', '-0.06415952742099762']), ('y', ['-0.06537354737520218', '-0.05939256399869919', '-0.05178237706422806', '-0.045250535011291504', '-0.03852912411093712', '-0.02902713045477867', '-0.0202875267714262', '-0.009306307882070541', '-0.0664256289601326', '-0.06074121594429016', '-0.053390298038721085', '-0.04606533423066139', '-0.03825720399618149', '-0.029871713370084763', '-0.019340993836522102', '-0.007975514978170395', '-0.06742905080318451', '-0.061004213988780975', '-0.05385209619998932', '-0.04655476287007332', '-0.03878721594810486', '-0.02883017435669899', '-0.017954347655177116', '-0.008002692833542824', '-0.06611424684524536', '-0.05987147241830826', '-0.05290590599179268', '-0.045647479593753815', '-0.03760911524295807', '-0.027332965284585953', '-0.017604675143957138', '-0.006250479258596897', '-0.06573858112096786', '-0.05929581820964813', '-0.05233205854892731', '-0.04382742941379547', '-0.035688526928424835', '-0.025391235947608948', '-0.015641164034605026', '-0.004400234669446945', '-0.063740074634552', '-0.057216327637434006', '-0.05005347728729248', '-0.0414251983165741', '-0.03339783847332001', '-0.02303282730281353', '-0.013502392917871475', '-0.0025412733666598797', '-0.06053349748253822', '-0.053560156375169754', '-0.047360196709632874', '-0.038510050624608994', '-0.03058759868144989', '-0.020398015156388283', '-0.011234592646360397', '-0.0006710337474942207', '-0.057449355721473694', '-0.049960020929574966', '-0.04335445538163185', '-0.03572570160031319', '-0.02766432613134384', '-0.017735280096530914', '-0.007426424417644739', '0.0015584935899823904']), ('z', ['0.07015674561262131', '0.07805224508047104', '0.08383159339427948', '0.0894855409860611', '0.093968465924263', '0.09850599616765976', '0.10092580318450928', '0.10348425060510635', '0.06912147253751755', '0.07691735774278641', '0.08223433047533035', '0.08709487318992615', '0.09175404906272888', '0.09489567577838898', '0.09894071519374847', '0.10255689918994904', '0.06775238364934921', '0.07377880066633224', '0.07930176705121994', '0.08423192799091339', '0.08779606223106384', '0.09262681752443314', '0.09548339992761612', '0.0977330431342125', '0.06322437524795532', '0.06903918087482452', '0.07436439394950867', '0.07952717691659927', '0.08295954763889313', '0.0881425067782402', '0.09012438356876373', '0.08837144076824188', '0.058145731687545776', '0.06322012841701508', '0.06939953565597534', '0.07313114404678345', '0.07694293558597565', '0.08083971589803696', '0.08229832351207733', '0.08078417181968689', '0.051265649497509', '0.05740974843502045', '0.06253481656312943', '0.06662151217460632', '0.07086562365293503', '0.07327373325824738', '0.07430699467658997', '0.073928602039814', '0.04550513997673988', '0.04998749867081642', '0.055407170206308365', '0.05841590464115143', '0.06297414004802704', '0.06437857449054718', '0.06384582072496414', '0.06401360034942627', '0.03716251999139786', '0.04119298979640007', '0.0462840236723423', '0.050298113375902176', '0.05298703908920288', '0.05405620113015175', '0.05301937460899353', '0.05387342721223831']), ('size', ['n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a'])])
/home/stefanappelhoff/Desktop/bids/mne-bids/mne_bids/read.py:221: RuntimeWarning: Fiducial point nasion not found, assuming identity unknown to head transformation
raw.set_montage(montage)
The channel coordinates after writing into BIDS:
[('POL LPG1-Ref', array([-0.01319475, -0.06537355, 0.07015675])),
('POL LPG2-Ref', array([-0.02327222, -0.06074122, 0.07691736])),
('POL LPG3-Ref', array([-0.02474644, -0.0533903 , 0.08223433])),
('POL LPG4-Ref', array([-0.02632337, -0.04606533, 0.08709487])),
('POL LPG5-Ref', array([-0.0261479 , -0.0382572 , 0.09175405]))]
<Figure size 640x640 with 1 Axes>
Total running time of the script: ( 0 minutes 0.168 seconds)