Tutorial on Computing HFOs (Part 3)¶
In this tutorial, we will walk through how to compare detectors. As in part 2, we will demonstrate this on a sample dataset that is defined in [1].
We will demonstrate compar of the following detectors:
Line Length detector
RMS detector
Morphology detector (used in the paper)
Dataset Preprocessing¶
Note that the data has been converted to BIDS to facilitate easy loading using mne-bids package. Another thing to note is that the authors in this dataset reported HFOs detected using bipolar montage. In addition, they only analyzed HFOs for a subset of the recording channels.
In order to compare results to a monopolar reference, we define an HFO to be “found” if there was an HFO in either of the corresponding bipolar contacts.
References¶
[1] Fedele T, Burnos S, Boran E, Krayenbühl N, Hilfiker P, Grunwald T, Sarnthein J. Resection of high frequency oscillations predicts seizure outcome in the individual patient. Scientific Reports. 2017;7(1):13836. https://www.nature.com/articles/s41598-017-13064-1 doi:10.1038/s41598-017-13064-1
[1]:
# first let's load in all our packages
import matplotlib
import matplotlib.pyplot as plt
import numpy as np
import sys
import os
import re
import pandas as pd
from mne_bids import (read_raw_bids, BIDSPath,
get_entity_vals, get_datatypes,
make_report)
from mne_bids.stats import count_events
import mne
from mne import make_ad_hoc_cov
basepath = os.path.join(os.getcwd(), "../..")
sys.path.append(basepath)
from mne_hfo import LineLengthDetector, RMSDetector
from mne_hfo.compare import compare_detectors
1 Working with Real Data¶
We are now going to work with the dataset from Fedele et al. linked above
1.1 Load in Real Data¶
We are going to skip intermediate/display steps since you did this in part 2
[2]:
%%capture
# this may change depending on where you store the data
root = "C:/Users/patri/Dropbox/fedele_hfo_data"
subjects = get_entity_vals(root, 'subject')
sessions = get_entity_vals(root, 'session')
subjectID = subjects[0]
sessionID = sessions[0]
bids_path = BIDSPath(subject=subjectID, session=sessionID,
datatype='ieeg',
suffix='ieeg',
extension='.vhdr', root=root)
# get first matching dataset
fpath = bids_path.match()[0]
# load dataset into mne Raw object
extra_params = dict(preload=True)
raw = read_raw_bids(fpath, extra_params)
1.2 Perform pre-processing steps¶
[3]:
def convert_to_bipolar(raw, drop_originals=True):
original_ch_names = raw.ch_names
ch_names_sorted = sorted(original_ch_names)
ch_pairs = []
for first, second in zip(ch_names_sorted, ch_names_sorted[1:]):
firstName = re.sub(r'[0-9]+', '', first)
secondName = re.sub(r'[0-9]+', '', second)
if firstName == secondName:
ch_pairs.append((first,second))
for ch_pair in ch_pairs:
raw = mne.set_bipolar_reference(raw, ch_pair[0], ch_pair[1], drop_refs=False)
if drop_originals:
raw = raw.drop_channels(original_ch_names)
return raw
[4]:
%%capture
raw = convert_to_bipolar(raw)
1.3 Perform Detection with Both Detectors¶
[5]:
# Set Key Word Arguments for the Line Length Detector and generate the class object
kwargs = {
'filter_band': (80, 250), # (l_freq, h_freq)
'threshold': 3, # Number of st. deviations
'win_size': 100, # Sliding window size in samples
'overlap': 0.25, # Fraction of window overlap [0, 1]
'hfo_name': "ripple"
}
ll_detector = LineLengthDetector(**kwargs)
# We use the same arguments for the RMS Detecor
rms_detector = RMSDetector(**kwargs)
[6]:
%%capture
# perform the fits
ll_detector = ll_detector.fit(raw)
rms_detector = rms_detector.fit(raw)
2 Compare detections¶
2.1 Perform the comparisons¶
We will calculate both the mutual information and cohen kappa scores to find agreement between detectors
[7]:
mutual_info = compare_detectors(ll_detector, rms_detector, method="mutual-info")
kappa = compare_detectors(ll_detector, rms_detector, method="cohen-kappa")
2.2 Visualize the comparisons¶
First let’s observe what the compare function returns
[8]:
mutual_info
[8]:
{'AHR1-AHR2': 0.006367438905637873,
'AHR2-AHR3': 0.009303055655412978,
'AHR3-AHR4': 0.008639619380071323,
'AHR4-AHR5': 0.040792175680735315,
'AHR5-AHR6': 0.016727304440477292,
'AHR6-AHR7': 0.008881367396859075,
'AHR7-AHR8': 0.006652660795102627,
'AL1-AL2': 0.12086053091711568,
'AL2-AL3': 0.30498226655255134,
'AL3-AL4': 0.3423289003563088,
'AL4-AL5': 0.35410834336008845,
'AL5-AL6': 0.2539488808327681,
'AL6-AL7': 0.27710634259216566,
'AL7-AL8': 0.20070048550752645,
'AR1-AR2': 0.009177030068343972,
'AR2-AR3': 0.02042497757073164,
'AR3-AR4': 0.1327554225272446,
'AR4-AR5': 0.16384897368786566,
'AR5-AR6': 0.21741452983633036,
'AR6-AR7': 0.14966156088411725,
'AR7-AR8': 0.029084598353392555,
'HL1-HL2': 0.02621331380334116,
'HL2-HL3': 0.0818495271081513,
'HL3-HL4': 0.08274279480227631,
'HL4-HL5': 0.03552038185412143,
'HL5-HL6': 0.36575250233949996,
'HL6-HL7': 0.26926841245696487,
'HL7-HL8': 0.2612247225578799,
'IAR1-IAR2': 0.2116504273602524,
'IAR2-IAR3': 0.09870475124445977,
'IAR3-IAR4': 0.11732768236785451,
'IAR4-IAR5': 0.0480649861738508,
'IAR5-IAR6': 0.0739228967961626,
'IPR1-IPR2': 0.21846108335962425,
'IPR2-IPR3': 0.16360431873072095,
'IPR3-IPR4': 0.09980904381865052,
'PHR1-PHR2': 0.0030805001333093635,
'PHR2-PHR3': 0.003443298440425263,
'PHR3-PHR4': 0.006087761645857337,
'PHR4-PHR5': 0.14717548817859138,
'PHR5-PHR6': 0.19563898293185206,
'PHR6-PHR7': 0.16490723866899876,
'PHR7-PHR8': 0.04689647206004438}
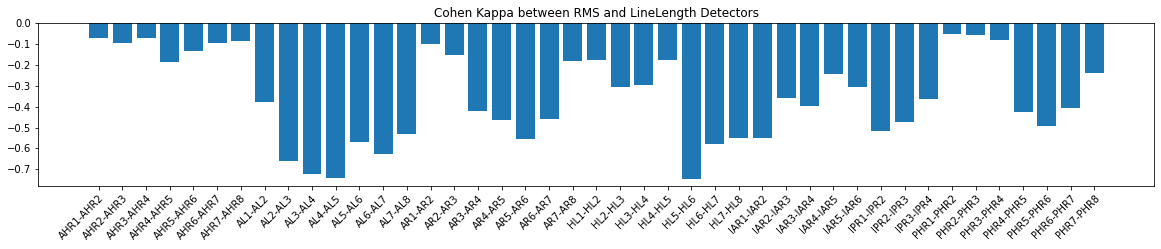
As you can see, the compare function gives a dictionary where the keys are the channels and the values are the metric value, in this case the mutual information. We can use these values to plot a barchart to display which channels the detections had the most agreement on.
2.1.1 Extract the right values¶
[9]:
ch_names = list(mutual_info.keys())
mutual_info_vals = list(mutual_info.values())
kappa_vals = list(kappa.values())
2.1.2 Visualize the mutual info¶
[10]:
fig, ax = plt.subplots(figsize=(20, 3))
bar = ax.bar(x=np.arange(len(ch_names)), height=mutual_info_vals)
ax.set_xticks(np.arange(len(ch_names)))
ax.set_xticklabels(ch_names)
plt.setp(ax.get_xticklabels(), rotation=45, ha="right", rotation_mode="anchor")
ax.set_title("Mutual info between RMS and LineLength Detectors")
#fig.tight_layout()
plt.show()
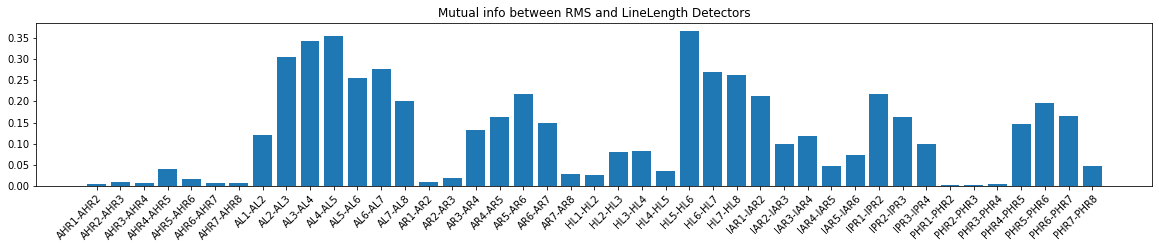
2.1.3 Visualize the Kappa score¶
[11]:
fig, ax = plt.subplots(figsize=(20, 3))
bar = ax.bar(x=np.arange(len(ch_names)), height=kappa_vals)
ax.set_xticks(np.arange(len(ch_names)))
ax.set_xticklabels(ch_names)
plt.setp(ax.get_xticklabels(), rotation=45, ha="right", rotation_mode="anchor")
ax.set_title("Cohen Kappa between RMS and LineLength Detectors")
#fig.tight_layout()
plt.show()