mne_nirs.statistics.ContrastResults#
- class mne_nirs.statistics.ContrastResults(info, data, design)[source]#
Class containing GLM contrast results.
- Parameters:
- Attributes:
ch_namesReturn the channel names.
compensation_gradeThe current gradient compensation grade.
- data
Methods
copy()Return a copy of the GLM results.
get_channel_types([picks, unique, only_data_chs])Get a list of channel type for each channel.
plot_topo([figsize, sphere])Plot topomap GLM contrast data.
save(fname[, overwrite])Save GLM results to disk.
scatter([conditions, exclude_no_interest, ...])Scatter plot of the GLM results.
to_dataframe([order])Return a tidy dataframe representing the GLM results.
- Returns:
- glm_estResultsGLM,
Result class.
- __contains__(ch_type)#
Check channel type membership.
- Parameters:
- ch_type
str Channel type to check for. Can be e.g.
'meg','eeg','stim', etc.
- ch_type
- Returns:
- inbool
Whether or not the instance contains the given channel type.
Examples
Channel type membership can be tested as:
>>> 'meg' in inst True >>> 'seeg' in inst False
- __hash__ = None#
- property compensation_grade#
The current gradient compensation grade.
- copy()#
Return a copy of the GLM results.
- Returns:
- instinstance of ResultsGLM
A copy of the object.
- get_channel_types(picks=None, unique=False, only_data_chs=False)#
Get a list of channel type for each channel.
- Parameters:
- picks
str| array_like |slice|None Channels to include. Slices and lists of integers will be interpreted as channel indices. In lists, channel type strings (e.g.,
['meg', 'eeg']) will pick channels of those types, channel name strings (e.g.,['MEG0111', 'MEG2623']will pick the given channels. Can also be the string values'all'to pick all channels, or'data'to pick data channels. None (default) will pick all channels. Bad channels are included by default. Note that channels ininfo['bads']will be included if their names or indices are explicitly provided.- uniquebool
Whether to return only unique channel types. Default is
False.- only_data_chsbool
Whether to ignore non-data channels. Default is
False.
- picks
- Returns:
- channel_types
list The channel types.
- channel_types
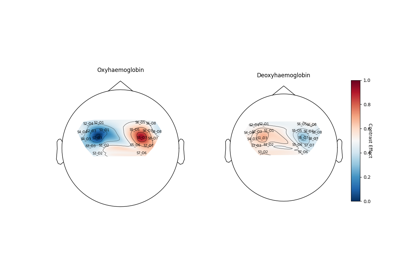
- plot_topo(figsize=(12, 7), sphere=None, **kwargs)[source]#
Plot topomap GLM contrast data.
- Parameters:
- figsize
numbers TODO: Remove this, how does MNE usually deal with this.
- sphere
numbers As specified in MNE.
- **kwargs
dict Additional keyword arguments to pass to
mne.viz.plot_topomap().Added in version 0.8.
- figsize
- Returns:
- figfigure
Figure of each design matrix component for hbo (top row) and hbr (bottom row).
- save(fname, overwrite=False)#
Save GLM results to disk.
- scatter(conditions=(), exclude_no_interest=True, axes=None, no_interest=None)#
Scatter plot of the GLM results.
- Parameters:
- conditions
list List of condition names to plot. By default plots all regressors of interest.
- exclude_no_interestbool
Exclude regressors of no interest from the figure.
- axesAxes
Optional axes on which to plot the data.
- no_interest
list List of regressors that are of no interest. If none are specified then conditions starting with [“drift”, “constant”, “short”, “Short”] will be excluded.
- conditions
- Returns:
- pltmatplotlib.Figure
Scatter plot.
- to_dataframe(order=None)#
Return a tidy dataframe representing the GLM results.
- Parameters:
- order
list Order in which the rows should be returned by channel name.
- order
- Returns:
- tidy
pandas.DataFrame Dataframe containing GLM results.
- tidy