Note
Click here to download the full example code
Reading/Writing a noise covariance matrix¶
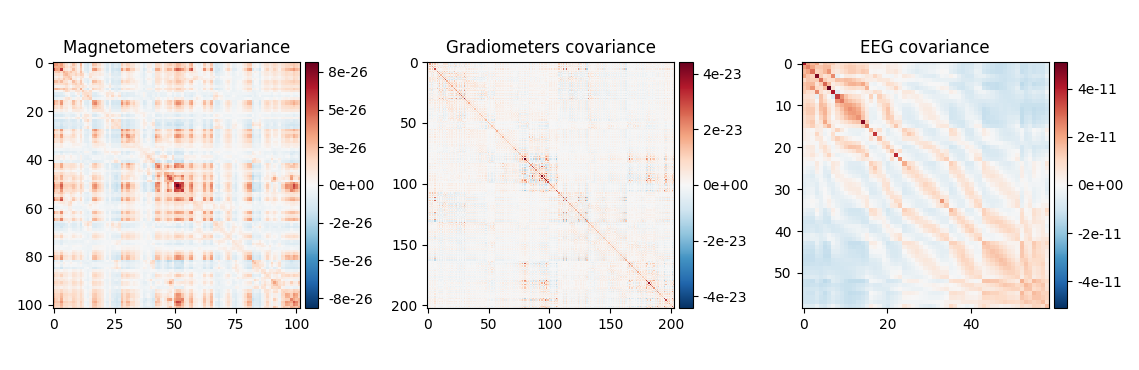
Plot a noise covariance matrix.
# Author: Alexandre Gramfort <alexandre.gramfort@inria.fr>
#
# License: BSD (3-clause)
from os import path as op
import mne
from mne.datasets import sample
print(__doc__)
data_path = sample.data_path()
fname_cov = op.join(data_path, 'MEG', 'sample', 'sample_audvis-cov.fif')
fname_evo = op.join(data_path, 'MEG', 'sample', 'sample_audvis-ave.fif')
cov = mne.read_cov(fname_cov)
print(cov)
evoked = mne.read_evokeds(fname_evo)[0]
Out:
366 x 366 full covariance (kind = 1) found.
Read a total of 4 projection items:
PCA-v1 (1 x 102) active
PCA-v2 (1 x 102) active
PCA-v3 (1 x 102) active
Average EEG reference (1 x 60) active
<Covariance | size : 366 x 366, n_samples : 15972, data : [[ 2.27235589e-23 4.79818505e-24 7.12852747e-25 ... 4.85348042e-18
2.02846360e-18 8.26727393e-18]
[ 4.79818505e-24 5.33468523e-24 1.80261790e-25 ... 2.33583009e-19
-6.93161055e-19 2.35199238e-18]
[ 7.12852747e-25 1.80261790e-25 5.79073915e-26 ... 1.09498615e-19
6.16924072e-21 2.93873875e-19]
...
[ 4.85348042e-18 2.33583009e-19 1.09498615e-19 ... 1.40677185e-11
1.27444183e-11 1.08634620e-11]
[ 2.02846360e-18 -6.93161055e-19 6.16924072e-21 ... 1.27444183e-11
1.59818134e-11 8.51070563e-12]
[ 8.26727393e-18 2.35199238e-18 2.93873875e-19 ... 1.08634620e-11
8.51070563e-12 1.53708918e-11]]>
Reading /home/circleci/mne_data/MNE-sample-data/MEG/sample/sample_audvis-ave.fif ...
Read a total of 4 projection items:
PCA-v1 (1 x 102) active
PCA-v2 (1 x 102) active
PCA-v3 (1 x 102) active
Average EEG reference (1 x 60) active
Found the data of interest:
t = -199.80 ... 499.49 ms (Left Auditory)
0 CTF compensation matrices available
nave = 55 - aspect type = 100
Projections have already been applied. Setting proj attribute to True.
No baseline correction applied
Read a total of 4 projection items:
PCA-v1 (1 x 102) active
PCA-v2 (1 x 102) active
PCA-v3 (1 x 102) active
Average EEG reference (1 x 60) active
Found the data of interest:
t = -199.80 ... 499.49 ms (Right Auditory)
0 CTF compensation matrices available
nave = 61 - aspect type = 100
Projections have already been applied. Setting proj attribute to True.
No baseline correction applied
Read a total of 4 projection items:
PCA-v1 (1 x 102) active
PCA-v2 (1 x 102) active
PCA-v3 (1 x 102) active
Average EEG reference (1 x 60) active
Found the data of interest:
t = -199.80 ... 499.49 ms (Left visual)
0 CTF compensation matrices available
nave = 67 - aspect type = 100
Projections have already been applied. Setting proj attribute to True.
No baseline correction applied
Read a total of 4 projection items:
PCA-v1 (1 x 102) active
PCA-v2 (1 x 102) active
PCA-v3 (1 x 102) active
Average EEG reference (1 x 60) active
Found the data of interest:
t = -199.80 ... 499.49 ms (Right visual)
0 CTF compensation matrices available
nave = 58 - aspect type = 100
Projections have already been applied. Setting proj attribute to True.
No baseline correction applied
Show covariance
cov.plot(evoked.info, exclude='bads', show_svd=False)
Total running time of the script: ( 0 minutes 2.775 seconds)
Estimated memory usage: 8 MB