Note
Click here to download the full example code
Plotting topographic arrowmaps of evoked data¶
Load evoked data and plot arrowmaps along with the topomap for selected time points. An arrowmap is based upon the Hosaka-Cohen transformation and represents an estimation of the current flow underneath the MEG sensors. They are a poor man’s MNE.
See 1 for details.
References¶
- 1
David Cohen and Hidehiro Hosaka. Part II magnetic field produced by a current dipole. Journal of Electrocardiology, 9(4):409–417, 1976. doi:10.1016/S0022-0736(76)80041-6.
# Authors: Sheraz Khan <sheraz@khansheraz.com>
#
# License: BSD (3-clause)
import numpy as np
import mne
from mne.datasets import sample
from mne.datasets.brainstorm import bst_raw
from mne import read_evokeds
from mne.viz import plot_arrowmap
print(__doc__)
path = sample.data_path()
fname = path + '/MEG/sample/sample_audvis-ave.fif'
# load evoked data
condition = 'Left Auditory'
evoked = read_evokeds(fname, condition=condition, baseline=(None, 0))
evoked_mag = evoked.copy().pick_types(meg='mag')
evoked_grad = evoked.copy().pick_types(meg='grad')
Out:
Reading /home/circleci/mne_data/MNE-sample-data/MEG/sample/sample_audvis-ave.fif ...
Read a total of 4 projection items:
PCA-v1 (1 x 102) active
PCA-v2 (1 x 102) active
PCA-v3 (1 x 102) active
Average EEG reference (1 x 60) active
Found the data of interest:
t = -199.80 ... 499.49 ms (Left Auditory)
0 CTF compensation matrices available
nave = 55 - aspect type = 100
Projections have already been applied. Setting proj attribute to True.
Applying baseline correction (mode: mean)
Removing projector <Projection | Average EEG reference, active : True, n_channels : 60>
Removing projector <Projection | PCA-v1, active : True, n_channels : 102>
Removing projector <Projection | PCA-v2, active : True, n_channels : 102>
Removing projector <Projection | PCA-v3, active : True, n_channels : 102>
Removing projector <Projection | Average EEG reference, active : True, n_channels : 60>
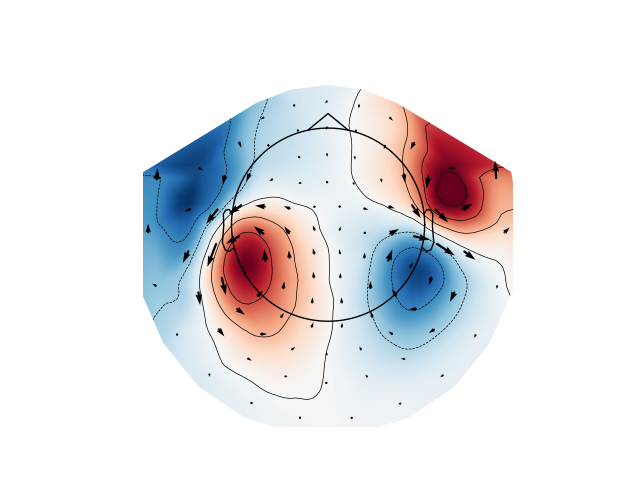
Plot magnetometer data as an arrowmap along with the topoplot at the time of the maximum sensor space activity:
max_time_idx = np.abs(evoked_mag.data).mean(axis=0).argmax()
plot_arrowmap(evoked_mag.data[:, max_time_idx], evoked_mag.info)
# Since planar gradiometers takes gradients along latitude and longitude,
# they need to be projected to the flatten manifold span by magnetometer
# or radial gradiometers before taking the gradients in the 2D Cartesian
# coordinate system for visualization on the 2D topoplot. You can use the
# ``info_from`` and ``info_to`` parameters to interpolate from
# gradiometer data to magnetometer data.
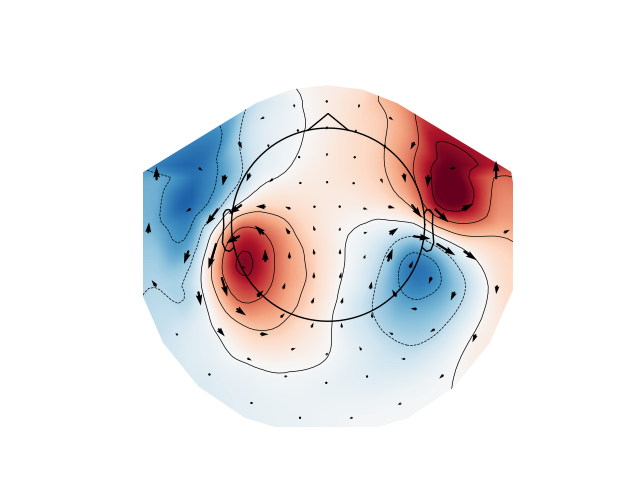
Plot gradiometer data as an arrowmap along with the topoplot at the time of the maximum sensor space activity:
plot_arrowmap(evoked_grad.data[:, max_time_idx], info_from=evoked_grad.info,
info_to=evoked_mag.info)
Out:
Computing dot products for 203 MEG channels...
Computing cross products for 203 → 102 MEG channels...
Preparing the mapping matrix...
Truncating at 79/203 components to omit less than 0.0001 (9.2e-05)
Since Vectorview 102 system perform sparse spatial sampling of the magnetic field, data from the Vectorview (info_from) can be projected to the high density CTF 272 system (info_to) for visualization
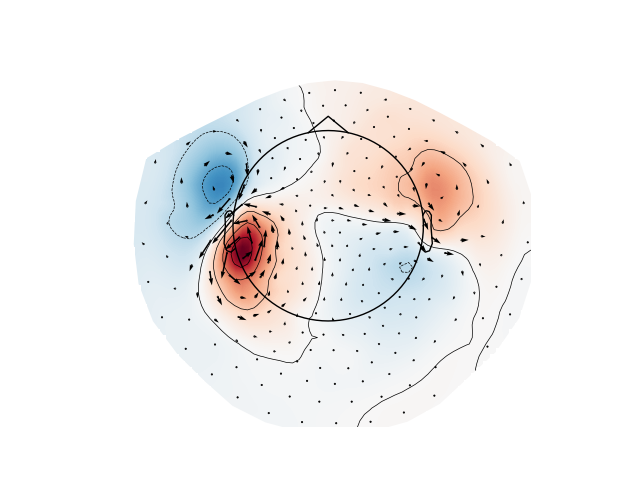
Plot gradiometer data as an arrowmap along with the topoplot at the time of the maximum sensor space activity:
path = bst_raw.data_path()
raw_fname = path + ('/MEG/bst_raw/'
'subj001_somatosensory_20111109_01_AUX-f.ds')
raw_ctf = mne.io.read_raw_ctf(raw_fname)
raw_ctf_info = mne.pick_info(
raw_ctf.info, mne.pick_types(raw_ctf.info, meg=True, ref_meg=False))
plot_arrowmap(evoked_grad.data[:, max_time_idx], info_from=evoked_grad.info,
info_to=raw_ctf_info, scale=6e-10)
Out:
ds directory : /home/circleci/mne_data/MNE-brainstorm-data/bst_raw/MEG/bst_raw/subj001_somatosensory_20111109_01_AUX-f.ds
res4 data read.
hc data read.
Separate EEG position data file not present.
Quaternion matching (desired vs. transformed):
0.84 69.49 0.00 mm <-> 0.84 69.49 -0.00 mm (orig : -44.30 51.45 -252.43 mm) diff = 0.000 mm
-0.84 -69.49 0.00 mm <-> -0.84 -69.49 -0.00 mm (orig : 46.28 -53.58 -243.47 mm) diff = 0.000 mm
86.41 0.00 0.00 mm <-> 86.41 0.00 0.00 mm (orig : 63.60 55.82 -230.26 mm) diff = 0.000 mm
Coordinate transformations established.
Reading digitizer points from ['/home/circleci/mne_data/MNE-brainstorm-data/bst_raw/MEG/bst_raw/subj001_somatosensory_20111109_01_AUX-f.ds/subj00111092011.pos']...
Polhemus data for 3 HPI coils added
Device coordinate locations for 3 HPI coils added
Picked positions of 2 EEG channels from channel info
2 EEG locations added to Polhemus data.
Measurement info composed.
Finding samples for /home/circleci/mne_data/MNE-brainstorm-data/bst_raw/MEG/bst_raw/subj001_somatosensory_20111109_01_AUX-f.ds/subj001_somatosensory_20111109_01_AUX-f.meg4:
System clock channel is available, checking which samples are valid.
240 x 1800 = 432000 samples from 302 chs
Current compensation grade : 3
Removing 5 compensators from info because not all compensation channels were picked.
Computing dot products for 203 MEG channels...
Computing cross products for 203 → 272 MEG channels...
Preparing the mapping matrix...
Truncating at 79/203 components to omit less than 0.0001 (9.2e-05)
Total running time of the script: ( 0 minutes 20.174 seconds)
Estimated memory usage: 9 MB