Roadmap¶
This page describes some of the major medium- to long-term goals for MNE-Python. These are goals that require substantial effort and/or API design considerations. Some of these may be suitable for Google Summer of Code projects, while others require more extensive work.
Page contents
Open¶
Time-frequency classes¶
Our current codebase implements classes related to TFRs that
remain incomplete. We should implement new classes from the ground up
that can hold frequency data (Spectrum), cross-spectral data
(CrossSpectrum), multitaper estimates (MultitaperSpectrum), and
time-varying estimates (Spectrogram). These should work for
continuous, epoched, and averaged sensor data, as well as source-space brain
data.
See related issues #6290, #7671, #8026, #8724, #9045, and PRs #6609, #6629, #6672, #6673, #8397, and #8892.
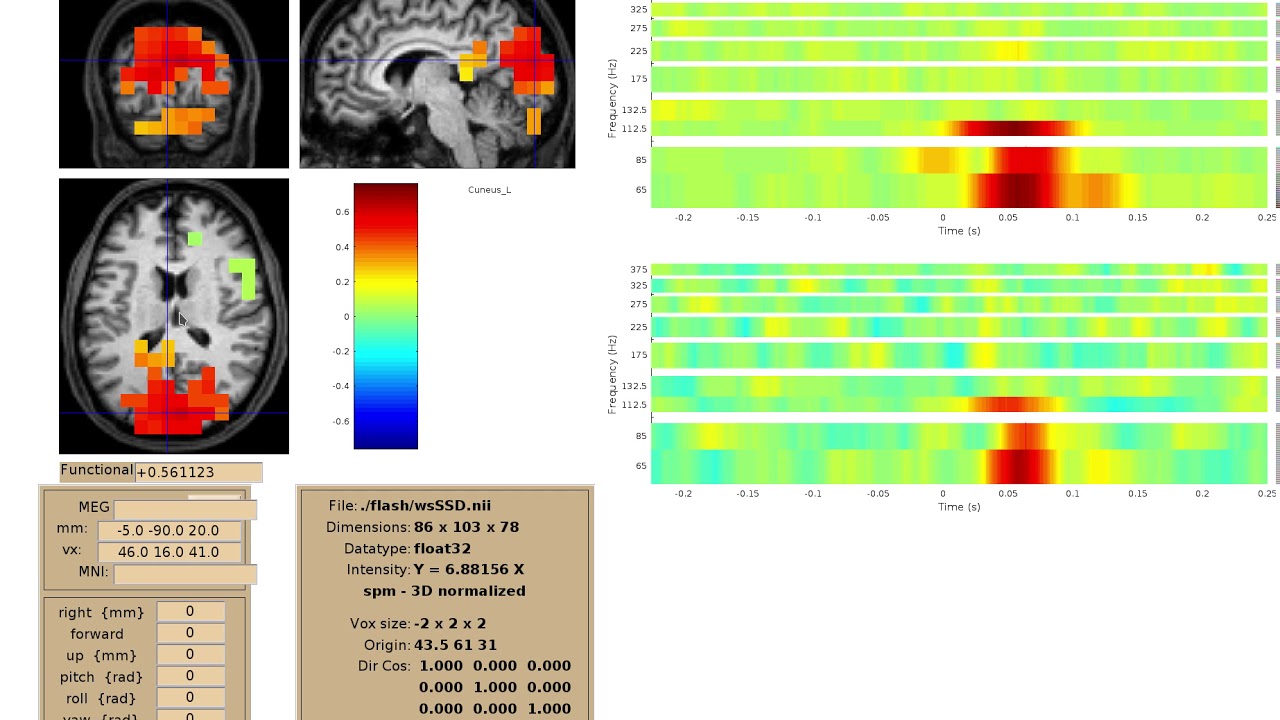
Time-frequency visualization¶
We should implement a viewer for interactive visualization of volumetric source-time-frequency (5-D) maps on MRI slices (orthogonal 2D viewer). NutmegTrip (written by Sarang Dalal) provides similar functionality in Matlab in conjunction with FieldTrip. Example of NutmegTrip’s source-time-frequency mode in action (click for link to YouTube):
First-class OPM support¶
MNE-Python has support for reading some OPM data formats such as FIF, but support is still rudimentary. Support should be added for other manufacturers, and standard (and/or novel) preprocessing routines should be added to deal with coregistration adjustment, forward modeling, and OPM-specific artifacts.
Better sEEG/ECoG/DBS support¶
Some support already exists for iEEG electrodes in MNE-Python thanks in part to standard abstractions. However, iEEG-specific pipeline steps (e.g., electrode localization) and visualizations (e.g., per-shaft topo plots, Time-frequency visualization) are missing. MNE-Python should work with members of the ECoG/sEEG community to work with or build in existing tools, and extend native functionality for depth electrodes.
Deep source modeling¶
Existing source modeling and inverse routines are not explicitly designed to deal with deep sources. Advanced algorithms exist from MGH for enhancing deep source localization, and these should be implemented and vetted in MNE-Python.
Clustering statistics API¶
The current clustering statistics code has limited functionality. It should be
re-worked to create a new cluster_based_statistic or similar function.
In particular, the new API should:
Support mixed within- and between-subjects designs, different statistical functions, etc. This should be done via a
designargument that mirrorspatsy.dmatrices()or similar community standard (e.g., this is what is used bystatsmodels.regression.linear_model.OLS).Have clear tutorials showing how different contrasts can be done (toy data).
Have clear tutorials showing some common analyses on real data (time-freq, sensor space, source space, etc.)
Not introduce any significant speed penalty (e.g., < 10% slower) compared to the existing, more specialized/limited functions.
More details are in #4859.
Access to open EEG/MEG databases¶
We should improve the access to open EEG/MEG databases via the
mne.datasets module, in other words improve our dataset fetchers.
We have physionet, but much more. Having a consistent API to access multiple
data sources would be great. See #2852 and #3585 for some ideas,
as well as:
- Human Connectome Project Datasets
Over a 3-year span (2012-2015), the Human Connectome Project (HCP) scanned 1,200 healthy adult subjects. The available data includes MR structural scans, behavioral data and (on a subset of the data) resting state and/or task MEG data.
- MMN dataset
Used for tutorial/publications applying DCM for ERP analysis using SPM.
- Kymata datasets
Current and archived EMEG measurement data, used to test hypotheses in the Kymata atlas. The participants are healthy human adults listening to the radio and/or watching films, and the data is comprised of (averaged) EEG and MEG sensor data and source current reconstructions.
- BNCI Horizon
BCI datasets.
Integrate OpenMEEG via improved Python bindings¶
OpenMEEG is a state-of-the art solver for forward modeling in the field of brain imaging with MEG/EEG. It solves numerically partial differential equations (PDE). It is written in C++ with Python bindings written in SWIG. The ambition of the project is to integrate OpenMEEG into MNE offering to MNE the ability to solve more forward problems (cortical mapping, intracranial recordings, etc.). Some software tasks that shall be completed:
Cleanup Python bindings (remove useless functions, check memory managements, etc.)
Write example scripts for OpenMEEG that automatically generate web pages as for MNE
Understand how MNE encodes info about sensors (location, orientation, integration points etc.) and allow OpenMEEG to be used.
Help package OpenMEEG for Debian/Ubuntu
Help manage the continuous integration system
In progress¶
Diversity, Equity, and Inclusion¶
MNE-Python is committed to recruiting and retaining a diverse pool of contributors, see #8221.
BIDS Integration¶
MNE-Python is in the process of providing automated analysis of BIDS-compliant datasets, see MNE-BIDS-Pipeline. Two key goals are:
Incorporating functionality from the mnefun pipeline, which has been used extensively for pediatric data analysis at I-LABS. Multiple processing steps (e.g., eSSS), sanity checks (e.g., cHPI quality), and reporting (e.g., SSP joint plots, SNR plots) should be ported over.
Adding support for cloud computing. Currently, cloud computing with M/EEG data requires multiple manual steps, including remote environment setup, data transfer, monitoring of remote jobs, and retrieval of output data/results. With the implementation in MNE-Docker, would be achieved in practice by:
One-time (or per-project) setup steps, setting up host keys, access tokens, etc.
In code, switch to cloud computing rather than local computing via a simple change of config parameters.
See also #6086.
Statistics efficiency¶
A key technique in functional neuroimaging analysis is clustering brain activity in adjacent regions prior to statistical analysis. An important clustering algorithm — threshold-free cluster enhancement (TFCE) — currently relies on computationally expensive permutations for hypothesis testing. A faster, probabilistic version of TFCE (pTFCE) is available, and we are in the process of implementing this new algorithm.
3D visualization¶
Historically we have used Mayavi for 3D visualization, but have faced limitations and challenges with it. We should work to use some other backend (e.g., PyVista) to get major improvements, such as:
Proper notebook support (through ipyvtklink) (complete)
Better interactivity with surface plots (complete)
Time-frequency plotting (complementary to volume-based Time-frequency visualization)
Integration of multiple functions as done in
mne_analyze, e.g., simultaneous source estimate viewing, field map viewing, head surface display, etc. These are all currently available in separate functions, but we should be able to combine them in a single plot as well.
The meta-issue for tracking to-do lists for surface plotting is #7162.
Coregistration / 3D viewer¶
mne coreg is an excellent tool for coregistration, but is limited by being tied to Mayavi, Traits, and TraitsUI. We should first refactor in several (mostly) separable steps:
Refactor code to use traitlets
GUI elements to use PyQt5 (rather than TraitsUI/pyface)
3D plotting to use our abstracted 3D viz functions rather than Mayavi
Refactor distance/fitting classes to public ones to enable the example from #6693.
Once this is done, we can effectively switch to a PyVista backend.
Documentation updates¶
Our documentation has many minor issues, which can be found under the tag #labels/DOC.
Completed¶
2D visualization¶
This goal was completed under CZI EOSS2. Some additional enhancements that could also be implemented are listed in #7751.
Tutorial / example overhaul¶
This goal was completed under CZI EOSS2. Ongoing documentation needs are listed in Documentation updates.
Cluster computing images¶
As part of this goal, we created docker images suitable for cloud computing via MNE-Docker. These will be integrated with MNE-BIDS-Pipeline to provide seamless cloud computing support of large datasets.