Note
Click here to download the full example code
Compute evoked ERS source power using DICS, LCMV beamformer, and dSPM¶
Here we examine 3 ways of localizing event-related synchronization (ERS) of beta band activity in this dataset: Somatosensory using DICS, LCMV beamformer, and dSPM applied to active and baseline covariance matrices.
# Authors: Luke Bloy <luke.bloy@gmail.com>
# Eric Larson <larson.eric.d@gmail.com>
#
# License: BSD (3-clause)
import os.path as op
import numpy as np
import mne
from mne.cov import compute_covariance
from mne.datasets import somato
from mne.time_frequency import csd_morlet
from mne.beamformer import (make_dics, apply_dics_csd, make_lcmv,
apply_lcmv_cov)
from mne.minimum_norm import (make_inverse_operator, apply_inverse_cov)
print(__doc__)
Reading the raw data and creating epochs:
data_path = somato.data_path()
subject = '01'
task = 'somato'
raw_fname = op.join(data_path, 'sub-{}'.format(subject), 'meg',
'sub-{}_task-{}_meg.fif'.format(subject, task))
raw = mne.io.read_raw_fif(raw_fname)
# We are interested in the beta band (12-30 Hz)
raw.load_data().filter(12, 30)
# The DICS beamformer currently only supports a single sensor type.
# We'll use the gradiometers in this example.
picks = mne.pick_types(raw.info, meg='grad', exclude='bads')
# Read epochs
events = mne.find_events(raw)
epochs = mne.Epochs(raw, events, event_id=1, tmin=-1.5, tmax=2, picks=picks,
preload=True)
# Read forward operator and point to freesurfer subject directory
fname_fwd = op.join(data_path, 'derivatives', 'sub-{}'.format(subject),
'sub-{}_task-{}-fwd.fif'.format(subject, task))
subjects_dir = op.join(data_path, 'derivatives', 'freesurfer', 'subjects')
fwd = mne.read_forward_solution(fname_fwd)
Out:
Opening raw data file /home/circleci/mne_data/MNE-somato-data/sub-01/meg/sub-01_task-somato_meg.fif...
Range : 237600 ... 506999 = 791.189 ... 1688.266 secs
Ready.
Reading 0 ... 269399 = 0.000 ... 897.077 secs...
Filtering raw data in 1 contiguous segment
Setting up band-pass filter from 12 - 30 Hz
FIR filter parameters
---------------------
Designing a one-pass, zero-phase, non-causal bandpass filter:
- Windowed time-domain design (firwin) method
- Hamming window with 0.0194 passband ripple and 53 dB stopband attenuation
- Lower passband edge: 12.00
- Lower transition bandwidth: 3.00 Hz (-6 dB cutoff frequency: 10.50 Hz)
- Upper passband edge: 30.00 Hz
- Upper transition bandwidth: 7.50 Hz (-6 dB cutoff frequency: 33.75 Hz)
- Filter length: 331 samples (1.102 sec)
111 events found
Event IDs: [1]
Not setting metadata
Not setting metadata
111 matching events found
Applying baseline correction (mode: mean)
0 projection items activated
Loading data for 111 events and 1052 original time points ...
0 bad epochs dropped
Reading forward solution from /home/circleci/mne_data/MNE-somato-data/derivatives/sub-01/sub-01_task-somato-fwd.fif...
Reading a source space...
[done]
Reading a source space...
[done]
2 source spaces read
Desired named matrix (kind = 3523) not available
Read MEG forward solution (8155 sources, 306 channels, free orientations)
Source spaces transformed to the forward solution coordinate frame
Compute covariances¶
ERS activity starts at 0.5 seconds after stimulus onset.
active_win = (0.5, 1.5)
baseline_win = (-1, 0)
baseline_cov = compute_covariance(epochs, tmin=baseline_win[0],
tmax=baseline_win[1], method='shrunk',
rank=None)
active_cov = compute_covariance(epochs, tmin=active_win[0], tmax=active_win[1],
method='shrunk', rank=None)
# Weighted averaging is already in the addition of covariance objects.
common_cov = baseline_cov + active_cov
Out:
Computing rank from data with rank=None
Using tolerance 1.9e-09 (2.2e-16 eps * 204 dim * 4.2e+04 max singular value)
Estimated rank (grad): 204
GRAD: rank 204 computed from 204 data channels with 0 projectors
Reducing data rank from 204 -> 204
Estimating covariance using SHRUNK
Done.
Number of samples used : 33411
[done]
Computing rank from data with rank=None
Using tolerance 2.1e-09 (2.2e-16 eps * 204 dim * 4.7e+04 max singular value)
Estimated rank (grad): 204
GRAD: rank 204 computed from 204 data channels with 0 projectors
Reducing data rank from 204 -> 204
Estimating covariance using SHRUNK
Done.
Number of samples used : 33411
[done]
Compute some source estimates¶
Here we will use DICS, LCMV beamformer, and dSPM.
See Compute source power using DICS beamformer for more information about DICS.
def _gen_dics(active_win, baseline_win, epochs):
freqs = np.logspace(np.log10(12), np.log10(30), 9)
csd = csd_morlet(epochs, freqs, tmin=-1, tmax=1.5, decim=20)
csd_baseline = csd_morlet(epochs, freqs, tmin=baseline_win[0],
tmax=baseline_win[1], decim=20)
csd_ers = csd_morlet(epochs, freqs, tmin=active_win[0], tmax=active_win[1],
decim=20)
filters = make_dics(epochs.info, fwd, csd.mean(), pick_ori='max-power')
stc_base, freqs = apply_dics_csd(csd_baseline.mean(), filters)
stc_act, freqs = apply_dics_csd(csd_ers.mean(), filters)
stc_act /= stc_base
return stc_act
# generate lcmv source estimate
def _gen_lcmv(active_cov, baseline_cov, common_cov):
filters = make_lcmv(epochs.info, fwd, common_cov, reg=0.05,
noise_cov=None, pick_ori='max-power')
stc_base = apply_lcmv_cov(baseline_cov, filters)
stc_act = apply_lcmv_cov(active_cov, filters)
stc_act /= stc_base
return stc_act
# generate mne/dSPM source estimate
def _gen_mne(active_cov, baseline_cov, common_cov, fwd, info, method='dSPM'):
inverse_operator = make_inverse_operator(info, fwd, common_cov)
stc_act = apply_inverse_cov(active_cov, info, inverse_operator,
method=method, verbose=True)
stc_base = apply_inverse_cov(baseline_cov, info, inverse_operator,
method=method, verbose=True)
stc_act /= stc_base
return stc_act
# Compute source estimates
stc_dics = _gen_dics(active_win, baseline_win, epochs)
stc_lcmv = _gen_lcmv(active_cov, baseline_cov, common_cov)
stc_dspm = _gen_mne(active_cov, baseline_cov, common_cov, fwd, epochs.info)
Out:
Computing cross-spectral density from epochs...
Computing CSD matrix for epoch 1
Computing CSD matrix for epoch 2
Computing CSD matrix for epoch 3
Computing CSD matrix for epoch 4
Computing CSD matrix for epoch 5
Computing CSD matrix for epoch 6
Computing CSD matrix for epoch 7
Computing CSD matrix for epoch 8
Computing CSD matrix for epoch 9
Computing CSD matrix for epoch 10
Computing CSD matrix for epoch 11
Computing CSD matrix for epoch 12
Computing CSD matrix for epoch 13
Computing CSD matrix for epoch 14
Computing CSD matrix for epoch 15
Computing CSD matrix for epoch 16
Computing CSD matrix for epoch 17
Computing CSD matrix for epoch 18
Computing CSD matrix for epoch 19
Computing CSD matrix for epoch 20
Computing CSD matrix for epoch 21
Computing CSD matrix for epoch 22
Computing CSD matrix for epoch 23
Computing CSD matrix for epoch 24
Computing CSD matrix for epoch 25
Computing CSD matrix for epoch 26
Computing CSD matrix for epoch 27
Computing CSD matrix for epoch 28
Computing CSD matrix for epoch 29
Computing CSD matrix for epoch 30
Computing CSD matrix for epoch 31
Computing CSD matrix for epoch 32
Computing CSD matrix for epoch 33
Computing CSD matrix for epoch 34
Computing CSD matrix for epoch 35
Computing CSD matrix for epoch 36
Computing CSD matrix for epoch 37
Computing CSD matrix for epoch 38
Computing CSD matrix for epoch 39
Computing CSD matrix for epoch 40
Computing CSD matrix for epoch 41
Computing CSD matrix for epoch 42
Computing CSD matrix for epoch 43
Computing CSD matrix for epoch 44
Computing CSD matrix for epoch 45
Computing CSD matrix for epoch 46
Computing CSD matrix for epoch 47
Computing CSD matrix for epoch 48
Computing CSD matrix for epoch 49
Computing CSD matrix for epoch 50
Computing CSD matrix for epoch 51
Computing CSD matrix for epoch 52
Computing CSD matrix for epoch 53
Computing CSD matrix for epoch 54
Computing CSD matrix for epoch 55
Computing CSD matrix for epoch 56
Computing CSD matrix for epoch 57
Computing CSD matrix for epoch 58
Computing CSD matrix for epoch 59
Computing CSD matrix for epoch 60
Computing CSD matrix for epoch 61
Computing CSD matrix for epoch 62
Computing CSD matrix for epoch 63
Computing CSD matrix for epoch 64
Computing CSD matrix for epoch 65
Computing CSD matrix for epoch 66
Computing CSD matrix for epoch 67
Computing CSD matrix for epoch 68
Computing CSD matrix for epoch 69
Computing CSD matrix for epoch 70
Computing CSD matrix for epoch 71
Computing CSD matrix for epoch 72
Computing CSD matrix for epoch 73
Computing CSD matrix for epoch 74
Computing CSD matrix for epoch 75
Computing CSD matrix for epoch 76
Computing CSD matrix for epoch 77
Computing CSD matrix for epoch 78
Computing CSD matrix for epoch 79
Computing CSD matrix for epoch 80
Computing CSD matrix for epoch 81
Computing CSD matrix for epoch 82
Computing CSD matrix for epoch 83
Computing CSD matrix for epoch 84
Computing CSD matrix for epoch 85
Computing CSD matrix for epoch 86
Computing CSD matrix for epoch 87
Computing CSD matrix for epoch 88
Computing CSD matrix for epoch 89
Computing CSD matrix for epoch 90
Computing CSD matrix for epoch 91
Computing CSD matrix for epoch 92
Computing CSD matrix for epoch 93
Computing CSD matrix for epoch 94
Computing CSD matrix for epoch 95
Computing CSD matrix for epoch 96
Computing CSD matrix for epoch 97
Computing CSD matrix for epoch 98
Computing CSD matrix for epoch 99
Computing CSD matrix for epoch 100
Computing CSD matrix for epoch 101
Computing CSD matrix for epoch 102
Computing CSD matrix for epoch 103
Computing CSD matrix for epoch 104
Computing CSD matrix for epoch 105
Computing CSD matrix for epoch 106
Computing CSD matrix for epoch 107
Computing CSD matrix for epoch 108
Computing CSD matrix for epoch 109
Computing CSD matrix for epoch 110
Computing CSD matrix for epoch 111
[done]
Computing cross-spectral density from epochs...
Computing CSD matrix for epoch 1
Computing CSD matrix for epoch 2
Computing CSD matrix for epoch 3
Computing CSD matrix for epoch 4
Computing CSD matrix for epoch 5
Computing CSD matrix for epoch 6
Computing CSD matrix for epoch 7
Computing CSD matrix for epoch 8
Computing CSD matrix for epoch 9
Computing CSD matrix for epoch 10
Computing CSD matrix for epoch 11
Computing CSD matrix for epoch 12
Computing CSD matrix for epoch 13
Computing CSD matrix for epoch 14
Computing CSD matrix for epoch 15
Computing CSD matrix for epoch 16
Computing CSD matrix for epoch 17
Computing CSD matrix for epoch 18
Computing CSD matrix for epoch 19
Computing CSD matrix for epoch 20
Computing CSD matrix for epoch 21
Computing CSD matrix for epoch 22
Computing CSD matrix for epoch 23
Computing CSD matrix for epoch 24
Computing CSD matrix for epoch 25
Computing CSD matrix for epoch 26
Computing CSD matrix for epoch 27
Computing CSD matrix for epoch 28
Computing CSD matrix for epoch 29
Computing CSD matrix for epoch 30
Computing CSD matrix for epoch 31
Computing CSD matrix for epoch 32
Computing CSD matrix for epoch 33
Computing CSD matrix for epoch 34
Computing CSD matrix for epoch 35
Computing CSD matrix for epoch 36
Computing CSD matrix for epoch 37
Computing CSD matrix for epoch 38
Computing CSD matrix for epoch 39
Computing CSD matrix for epoch 40
Computing CSD matrix for epoch 41
Computing CSD matrix for epoch 42
Computing CSD matrix for epoch 43
Computing CSD matrix for epoch 44
Computing CSD matrix for epoch 45
Computing CSD matrix for epoch 46
Computing CSD matrix for epoch 47
Computing CSD matrix for epoch 48
Computing CSD matrix for epoch 49
Computing CSD matrix for epoch 50
Computing CSD matrix for epoch 51
Computing CSD matrix for epoch 52
Computing CSD matrix for epoch 53
Computing CSD matrix for epoch 54
Computing CSD matrix for epoch 55
Computing CSD matrix for epoch 56
Computing CSD matrix for epoch 57
Computing CSD matrix for epoch 58
Computing CSD matrix for epoch 59
Computing CSD matrix for epoch 60
Computing CSD matrix for epoch 61
Computing CSD matrix for epoch 62
Computing CSD matrix for epoch 63
Computing CSD matrix for epoch 64
Computing CSD matrix for epoch 65
Computing CSD matrix for epoch 66
Computing CSD matrix for epoch 67
Computing CSD matrix for epoch 68
Computing CSD matrix for epoch 69
Computing CSD matrix for epoch 70
Computing CSD matrix for epoch 71
Computing CSD matrix for epoch 72
Computing CSD matrix for epoch 73
Computing CSD matrix for epoch 74
Computing CSD matrix for epoch 75
Computing CSD matrix for epoch 76
Computing CSD matrix for epoch 77
Computing CSD matrix for epoch 78
Computing CSD matrix for epoch 79
Computing CSD matrix for epoch 80
Computing CSD matrix for epoch 81
Computing CSD matrix for epoch 82
Computing CSD matrix for epoch 83
Computing CSD matrix for epoch 84
Computing CSD matrix for epoch 85
Computing CSD matrix for epoch 86
Computing CSD matrix for epoch 87
Computing CSD matrix for epoch 88
Computing CSD matrix for epoch 89
Computing CSD matrix for epoch 90
Computing CSD matrix for epoch 91
Computing CSD matrix for epoch 92
Computing CSD matrix for epoch 93
Computing CSD matrix for epoch 94
Computing CSD matrix for epoch 95
Computing CSD matrix for epoch 96
Computing CSD matrix for epoch 97
Computing CSD matrix for epoch 98
Computing CSD matrix for epoch 99
Computing CSD matrix for epoch 100
Computing CSD matrix for epoch 101
Computing CSD matrix for epoch 102
Computing CSD matrix for epoch 103
Computing CSD matrix for epoch 104
Computing CSD matrix for epoch 105
Computing CSD matrix for epoch 106
Computing CSD matrix for epoch 107
Computing CSD matrix for epoch 108
Computing CSD matrix for epoch 109
Computing CSD matrix for epoch 110
Computing CSD matrix for epoch 111
[done]
Computing cross-spectral density from epochs...
Computing CSD matrix for epoch 1
Computing CSD matrix for epoch 2
Computing CSD matrix for epoch 3
Computing CSD matrix for epoch 4
Computing CSD matrix for epoch 5
Computing CSD matrix for epoch 6
Computing CSD matrix for epoch 7
Computing CSD matrix for epoch 8
Computing CSD matrix for epoch 9
Computing CSD matrix for epoch 10
Computing CSD matrix for epoch 11
Computing CSD matrix for epoch 12
Computing CSD matrix for epoch 13
Computing CSD matrix for epoch 14
Computing CSD matrix for epoch 15
Computing CSD matrix for epoch 16
Computing CSD matrix for epoch 17
Computing CSD matrix for epoch 18
Computing CSD matrix for epoch 19
Computing CSD matrix for epoch 20
Computing CSD matrix for epoch 21
Computing CSD matrix for epoch 22
Computing CSD matrix for epoch 23
Computing CSD matrix for epoch 24
Computing CSD matrix for epoch 25
Computing CSD matrix for epoch 26
Computing CSD matrix for epoch 27
Computing CSD matrix for epoch 28
Computing CSD matrix for epoch 29
Computing CSD matrix for epoch 30
Computing CSD matrix for epoch 31
Computing CSD matrix for epoch 32
Computing CSD matrix for epoch 33
Computing CSD matrix for epoch 34
Computing CSD matrix for epoch 35
Computing CSD matrix for epoch 36
Computing CSD matrix for epoch 37
Computing CSD matrix for epoch 38
Computing CSD matrix for epoch 39
Computing CSD matrix for epoch 40
Computing CSD matrix for epoch 41
Computing CSD matrix for epoch 42
Computing CSD matrix for epoch 43
Computing CSD matrix for epoch 44
Computing CSD matrix for epoch 45
Computing CSD matrix for epoch 46
Computing CSD matrix for epoch 47
Computing CSD matrix for epoch 48
Computing CSD matrix for epoch 49
Computing CSD matrix for epoch 50
Computing CSD matrix for epoch 51
Computing CSD matrix for epoch 52
Computing CSD matrix for epoch 53
Computing CSD matrix for epoch 54
Computing CSD matrix for epoch 55
Computing CSD matrix for epoch 56
Computing CSD matrix for epoch 57
Computing CSD matrix for epoch 58
Computing CSD matrix for epoch 59
Computing CSD matrix for epoch 60
Computing CSD matrix for epoch 61
Computing CSD matrix for epoch 62
Computing CSD matrix for epoch 63
Computing CSD matrix for epoch 64
Computing CSD matrix for epoch 65
Computing CSD matrix for epoch 66
Computing CSD matrix for epoch 67
Computing CSD matrix for epoch 68
Computing CSD matrix for epoch 69
Computing CSD matrix for epoch 70
Computing CSD matrix for epoch 71
Computing CSD matrix for epoch 72
Computing CSD matrix for epoch 73
Computing CSD matrix for epoch 74
Computing CSD matrix for epoch 75
Computing CSD matrix for epoch 76
Computing CSD matrix for epoch 77
Computing CSD matrix for epoch 78
Computing CSD matrix for epoch 79
Computing CSD matrix for epoch 80
Computing CSD matrix for epoch 81
Computing CSD matrix for epoch 82
Computing CSD matrix for epoch 83
Computing CSD matrix for epoch 84
Computing CSD matrix for epoch 85
Computing CSD matrix for epoch 86
Computing CSD matrix for epoch 87
Computing CSD matrix for epoch 88
Computing CSD matrix for epoch 89
Computing CSD matrix for epoch 90
Computing CSD matrix for epoch 91
Computing CSD matrix for epoch 92
Computing CSD matrix for epoch 93
Computing CSD matrix for epoch 94
Computing CSD matrix for epoch 95
Computing CSD matrix for epoch 96
Computing CSD matrix for epoch 97
Computing CSD matrix for epoch 98
Computing CSD matrix for epoch 99
Computing CSD matrix for epoch 100
Computing CSD matrix for epoch 101
Computing CSD matrix for epoch 102
Computing CSD matrix for epoch 103
Computing CSD matrix for epoch 104
Computing CSD matrix for epoch 105
Computing CSD matrix for epoch 106
Computing CSD matrix for epoch 107
Computing CSD matrix for epoch 108
Computing CSD matrix for epoch 109
Computing CSD matrix for epoch 110
Computing CSD matrix for epoch 111
[done]
Identifying common channels ...
Dropped the following channels:
['MEG 1211', 'MEG 2641', 'MEG 0531', 'MEG 0911', 'MEG 0931', 'MEG 2041', 'MEG 1111', 'MEG 1921', 'MEG 2231', 'MEG 1511', 'MEG 1221', 'MEG 1531', 'MEG 2011', 'MEG 1721', 'MEG 0821', 'MEG 1731', 'MEG 0121', 'MEG 1121', 'MEG 0411', 'MEG 2141', 'MEG 0241', 'MEG 2631', 'MEG 1611', 'MEG 1931', 'MEG 2431', 'MEG 0611', 'MEG 2311', 'MEG 0131', 'MEG 1831', 'MEG 1421', 'MEG 0941', 'MEG 2321', 'MEG 1741', 'MEG 0421', 'MEG 1431', 'MEG 2421', 'MEG 1241', 'MEG 2211', 'MEG 2531', 'MEG 1711', 'MEG 2611', 'MEG 0711', 'MEG 2441', 'MEG 1941', 'MEG 0441', 'MEG 1541', 'MEG 0331', 'MEG 0431', 'MEG 1811', 'MEG 0221', 'MEG 2341', 'MEG 0541', 'MEG 0321', 'MEG 0741', 'MEG 2331', 'MEG 0211', 'MEG 2541', 'MEG 1841', 'MEG 0511', 'MEG 0341', 'MEG 1821', 'MEG 1621', 'MEG 0521', 'MEG 1641', 'MEG 0921', 'MEG 2221', 'MEG 1411', 'MEG 0631', 'MEG 0141', 'MEG 2131', 'MEG 1141', 'MEG 1341', 'MEG 2521', 'MEG 0731', 'MEG 1021', 'MEG 1911', 'MEG 1521', 'MEG 2511', 'MEG 1031', 'MEG 1011', 'MEG 0111', 'MEG 0721', 'MEG 1331', 'MEG 2031', 'MEG 2411', 'MEG 1231', 'MEG 2621', 'MEG 1321', 'MEG 0811', 'MEG 1441', 'MEG 1041', 'MEG 1311', 'MEG 0231', 'MEG 0621', 'MEG 0311', 'MEG 1631', 'MEG 1131', 'MEG 2111', 'MEG 2121', 'MEG 0641', 'MEG 2241', 'MEG 2021']
Computing inverse operator with 204 channels.
204 out of 306 channels remain after picking
Selected 204 channels
Creating the depth weighting matrix...
Whitening the forward solution.
Computing rank from covariance with rank=None
Using tolerance 4.5e+08 (2.2e-16 eps * 204 dim * 1e+22 max singular value)
Estimated rank (grad): 204
GRAD: rank 204 computed from 204 data channels with 0 projectors
Setting small GRAD eigenvalues to zero (without PCA)
Creating the source covariance matrix
Adjusting source covariance matrix.
Computing DICS spatial filters...
Computing beamformer filters for 8155 sources
Filter computation complete
Computing DICS source power...
[done]
Computing DICS source power...
[done]
Computing rank from covariance with rank='info'
GRAD: rank 204 after 0 projectors applied to 204 channels
Computing rank from covariance with rank='info'
GRAD: rank 204 after 0 projectors applied to 204 channels
Making LCMV beamformer with rank {'grad': 204}
Computing inverse operator with 204 channels.
204 out of 306 channels remain after picking
Selected 204 channels
Whitening the forward solution.
Computing rank from covariance with rank={'grad': 204}
Setting small GRAD eigenvalues to zero (without PCA)
Creating the source covariance matrix
Adjusting source covariance matrix.
Computing beamformer filters for 8155 sources
Filter computation complete
Converting forward solution to surface orientation
No patch info available. The standard source space normals will be employed in the rotation to the local surface coordinates....
Converting to surface-based source orientations...
[done]
Computing inverse operator with 204 channels.
204 out of 306 channels remain after picking
Selected 204 channels
Creating the depth weighting matrix...
204 planar channels
limit = 7615/8155 = 10.004172
scale = 5.17919e-08 exp = 0.8
Applying loose dipole orientations to surface source spaces: 0.2
Whitening the forward solution.
Computing rank from covariance with rank=None
Using tolerance 2.6e-13 (2.2e-16 eps * 204 dim * 5.7 max singular value)
Estimated rank (grad): 204
GRAD: rank 204 computed from 204 data channels with 0 projectors
Setting small GRAD eigenvalues to zero (without PCA)
Creating the source covariance matrix
Adjusting source covariance matrix.
Computing SVD of whitened and weighted lead field matrix.
largest singular value = 4.31582
scaling factor to adjust the trace = 2.53127e+21
Preparing the inverse operator for use...
Scaled noise and source covariance from nave = 1 to nave = 1
Created the regularized inverter
The projection vectors do not apply to these channels.
Created the whitener using a noise covariance matrix with rank 204 (0 small eigenvalues omitted)
Computing noise-normalization factors (dSPM)...
[done]
Applying inverse operator to "cov"...
Picked 204 channels from the data
Computing inverse...
Eigenleads need to be weighted ...
Computing residual...
Explained 56.5% variance
dSPM...
Combining the current components...
[done]
Preparing the inverse operator for use...
Scaled noise and source covariance from nave = 1 to nave = 1
Created the regularized inverter
The projection vectors do not apply to these channels.
Created the whitener using a noise covariance matrix with rank 204 (0 small eigenvalues omitted)
Computing noise-normalization factors (dSPM)...
[done]
Applying inverse operator to "cov"...
Picked 204 channels from the data
Computing inverse...
Eigenleads need to be weighted ...
Computing residual...
Explained 56.5% variance
dSPM...
Combining the current components...
[done]
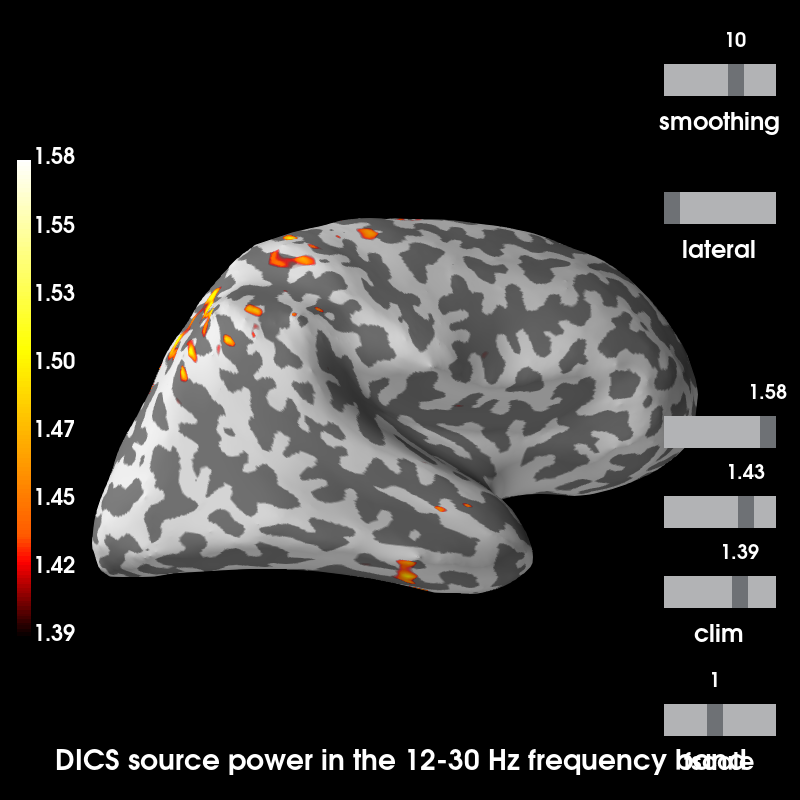
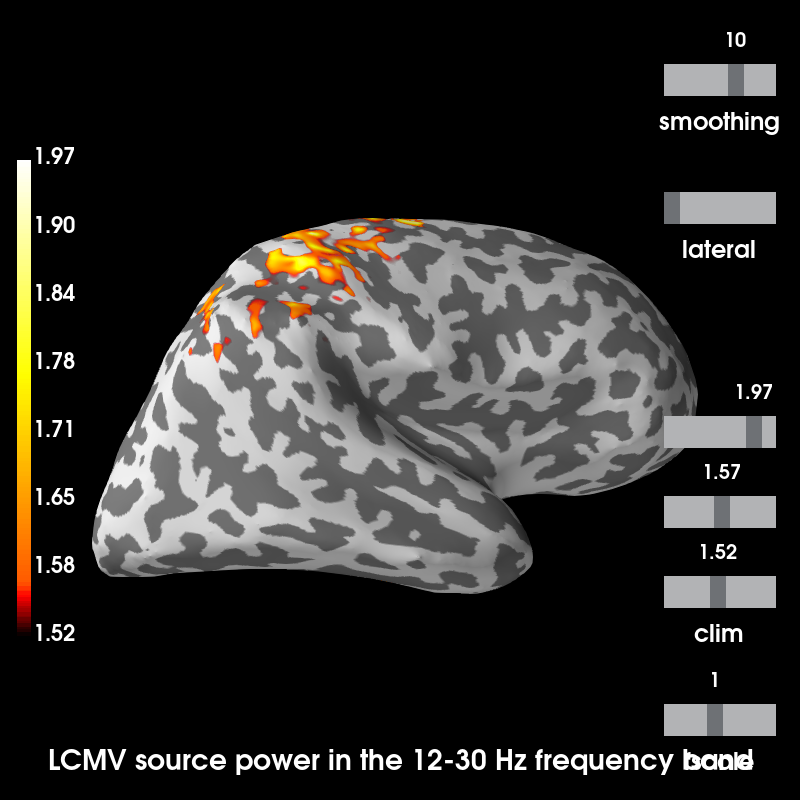
Plot source estimates¶
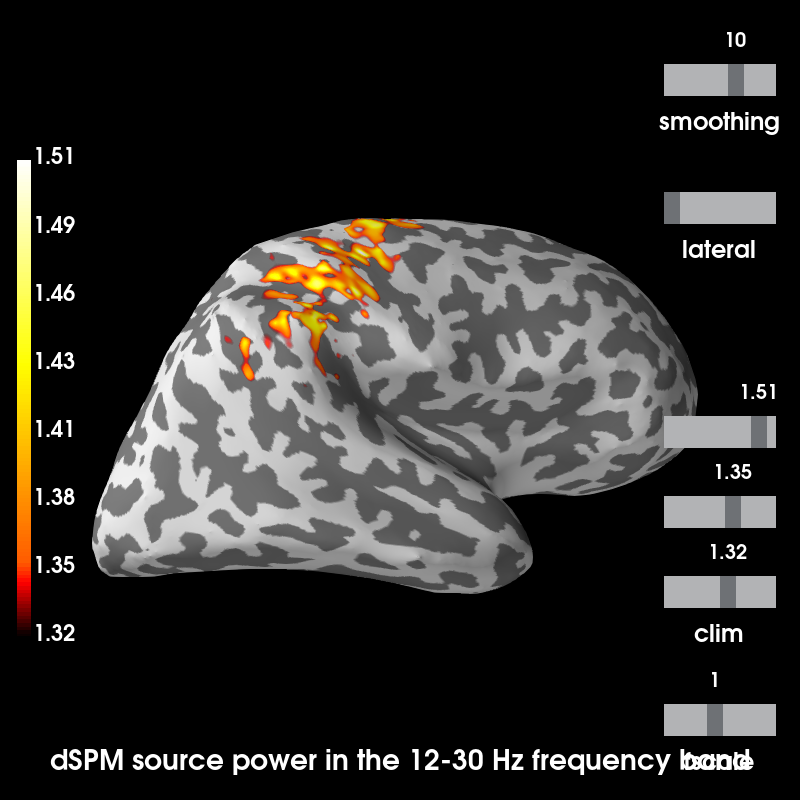
Out:
Using control points [1.39376309 1.43246371 1.57854484]
Using control points [1.51924176 1.56913499 1.96717505]
Using control points [1.32360332 1.35265114 1.51367404]
Total running time of the script: ( 0 minutes 59.773 seconds)
Estimated memory usage: 1202 MB