Note
Click here to download the full example code
Compute seed-based time-frequency connectivity in sensor space¶
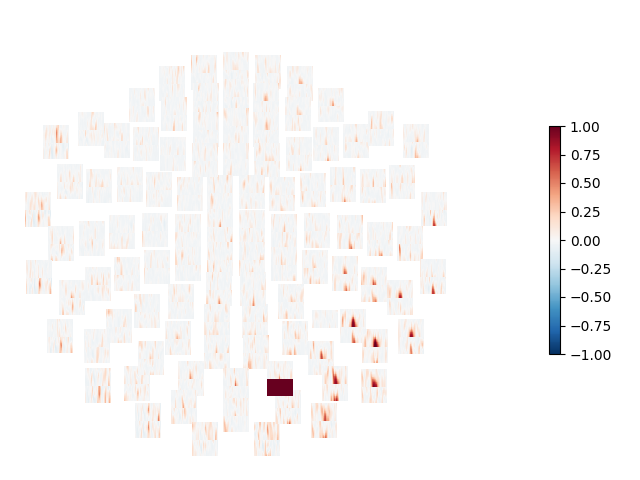
Computes the connectivity between a seed-gradiometer close to the visual cortex and all other gradiometers. The connectivity is computed in the time-frequency domain using Morlet wavelets and the debiased squared weighted phase lag index 1 is used as connectivity metric.
# Author: Martin Luessi <mluessi@nmr.mgh.harvard.edu>
#
# License: BSD (3-clause)
import numpy as np
import mne
from mne import io
from mne.connectivity import spectral_connectivity, seed_target_indices
from mne.datasets import sample
from mne.time_frequency import AverageTFR
print(__doc__)
Set parameters
data_path = sample.data_path()
raw_fname = data_path + '/MEG/sample/sample_audvis_filt-0-40_raw.fif'
event_fname = data_path + '/MEG/sample/sample_audvis_filt-0-40_raw-eve.fif'
# Setup for reading the raw data
raw = io.read_raw_fif(raw_fname)
events = mne.read_events(event_fname)
# Add a bad channel
raw.info['bads'] += ['MEG 2443']
# Pick MEG gradiometers
picks = mne.pick_types(raw.info, meg='grad', eeg=False, stim=False, eog=True,
exclude='bads')
# Create epochs for left-visual condition
event_id, tmin, tmax = 3, -0.2, 0.5
epochs = mne.Epochs(raw, events, event_id, tmin, tmax, picks=picks,
baseline=(None, 0), reject=dict(grad=4000e-13, eog=150e-6),
preload=True)
# Use 'MEG 2343' as seed
seed_ch = 'MEG 2343'
picks_ch_names = [raw.ch_names[i] for i in picks]
# Create seed-target indices for connectivity computation
seed = picks_ch_names.index(seed_ch)
targets = np.arange(len(picks))
indices = seed_target_indices(seed, targets)
# Define wavelet frequencies and number of cycles
cwt_freqs = np.arange(7, 30, 2)
cwt_n_cycles = cwt_freqs / 7.
# Run the connectivity analysis using 2 parallel jobs
sfreq = raw.info['sfreq'] # the sampling frequency
con, freqs, times, _, _ = spectral_connectivity(
epochs, indices=indices,
method='wpli2_debiased', mode='cwt_morlet', sfreq=sfreq,
cwt_freqs=cwt_freqs, cwt_n_cycles=cwt_n_cycles, n_jobs=1)
# Mark the seed channel with a value of 1.0, so we can see it in the plot
con[np.where(indices[1] == seed)] = 1.0
# Show topography of connectivity from seed
title = 'WPLI2 - Visual - Seed %s' % seed_ch
layout = mne.find_layout(epochs.info, 'meg') # use full layout
tfr = AverageTFR(epochs.info, con, times, freqs, len(epochs))
tfr.plot_topo(fig_facecolor='w', font_color='k', border='k')
Out:
Opening raw data file /home/circleci/mne_data/MNE-sample-data/MEG/sample/sample_audvis_filt-0-40_raw.fif...
Read a total of 4 projection items:
PCA-v1 (1 x 102) idle
PCA-v2 (1 x 102) idle
PCA-v3 (1 x 102) idle
Average EEG reference (1 x 60) idle
Range : 6450 ... 48149 = 42.956 ... 320.665 secs
Ready.
Not setting metadata
Not setting metadata
73 matching events found
Setting baseline interval to [-0.19979521315838786, 0.0] sec
Applying baseline correction (mode: mean)
4 projection items activated
Loading data for 73 events and 106 original time points ...
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
6 bad epochs dropped
Connectivity computation...
computing connectivity for 204 connections
using t=0.000s..0.699s for estimation (106 points)
frequencies: 9.0Hz..29.0Hz (11 points)
using CWT with Morlet wavelets to estimate spectra
the following metrics will be computed: Debiased WPLI Square
computing connectivity for epoch 1
computing connectivity for epoch 2
computing connectivity for epoch 3
computing connectivity for epoch 4
computing connectivity for epoch 5
computing connectivity for epoch 6
computing connectivity for epoch 7
computing connectivity for epoch 8
computing connectivity for epoch 9
computing connectivity for epoch 10
computing connectivity for epoch 11
computing connectivity for epoch 12
computing connectivity for epoch 13
computing connectivity for epoch 14
computing connectivity for epoch 15
computing connectivity for epoch 16
computing connectivity for epoch 17
computing connectivity for epoch 18
computing connectivity for epoch 19
computing connectivity for epoch 20
computing connectivity for epoch 21
computing connectivity for epoch 22
computing connectivity for epoch 23
computing connectivity for epoch 24
computing connectivity for epoch 25
computing connectivity for epoch 26
computing connectivity for epoch 27
computing connectivity for epoch 28
computing connectivity for epoch 29
computing connectivity for epoch 30
computing connectivity for epoch 31
computing connectivity for epoch 32
computing connectivity for epoch 33
computing connectivity for epoch 34
computing connectivity for epoch 35
computing connectivity for epoch 36
computing connectivity for epoch 37
computing connectivity for epoch 38
computing connectivity for epoch 39
computing connectivity for epoch 40
computing connectivity for epoch 41
computing connectivity for epoch 42
computing connectivity for epoch 43
computing connectivity for epoch 44
computing connectivity for epoch 45
computing connectivity for epoch 46
computing connectivity for epoch 47
computing connectivity for epoch 48
computing connectivity for epoch 49
computing connectivity for epoch 50
computing connectivity for epoch 51
computing connectivity for epoch 52
computing connectivity for epoch 53
computing connectivity for epoch 54
computing connectivity for epoch 55
computing connectivity for epoch 56
computing connectivity for epoch 57
computing connectivity for epoch 58
computing connectivity for epoch 59
computing connectivity for epoch 60
computing connectivity for epoch 61
computing connectivity for epoch 62
computing connectivity for epoch 63
computing connectivity for epoch 64
computing connectivity for epoch 65
computing connectivity for epoch 66
computing connectivity for epoch 67
[Connectivity computation done]
No baseline correction applied
References¶
- 1
Martin Vinck, Robert Oostenveld, Marijn van Wingerden, Franscesco Battaglia, and Cyriel M.A. Pennartz. An improved index of phase-synchronization for electrophysiological data in the presence of volume-conduction, noise and sample-size bias. NeuroImage, 55(4):1548–1565, 2011. doi:10.1016/j.neuroimage.2011.01.055.
Total running time of the script: ( 0 minutes 6.920 seconds)
Estimated memory usage: 8 MB