Note
Click here to download the full example code
Compute ICA components on epochs¶
ICA is fit to MEG raw data. We assume that the non-stationary EOG artifacts have already been removed. The sources matching the ECG are automatically found and displayed.
Note
This example does quite a bit of processing, so even on a fast machine it can take about a minute to complete.
# Authors: Denis Engemann <denis.engemann@gmail.com>
#
# License: BSD (3-clause)
import mne
from mne.preprocessing import ICA, create_ecg_epochs
from mne.datasets import sample
print(__doc__)
Read and preprocess the data. Preprocessing consists of:
MEG channel selection
1-30 Hz band-pass filter
epoching -0.2 to 0.5 seconds with respect to events
rejection based on peak-to-peak amplitude
data_path = sample.data_path()
raw_fname = data_path + '/MEG/sample/sample_audvis_filt-0-40_raw.fif'
raw = mne.io.read_raw_fif(raw_fname)
raw.pick_types(meg=True, eeg=False, exclude='bads', stim=True).load_data()
raw.filter(1, 30, fir_design='firwin')
# peak-to-peak amplitude rejection parameters
reject = dict(grad=4000e-13, mag=4e-12)
# longer + more epochs for more artifact exposure
events = mne.find_events(raw, stim_channel='STI 014')
epochs = mne.Epochs(raw, events, event_id=None, tmin=-0.2, tmax=0.5,
reject=reject)
Out:
Opening raw data file /home/circleci/mne_data/MNE-sample-data/MEG/sample/sample_audvis_filt-0-40_raw.fif...
Read a total of 4 projection items:
PCA-v1 (1 x 102) idle
PCA-v2 (1 x 102) idle
PCA-v3 (1 x 102) idle
Average EEG reference (1 x 60) idle
Range : 6450 ... 48149 = 42.956 ... 320.665 secs
Ready.
Removing projector <Projection | Average EEG reference, active : False, n_channels : 60>
Reading 0 ... 41699 = 0.000 ... 277.709 secs...
Filtering raw data in 1 contiguous segment
Setting up band-pass filter from 1 - 30 Hz
FIR filter parameters
---------------------
Designing a one-pass, zero-phase, non-causal bandpass filter:
- Windowed time-domain design (firwin) method
- Hamming window with 0.0194 passband ripple and 53 dB stopband attenuation
- Lower passband edge: 1.00
- Lower transition bandwidth: 1.00 Hz (-6 dB cutoff frequency: 0.50 Hz)
- Upper passband edge: 30.00 Hz
- Upper transition bandwidth: 7.50 Hz (-6 dB cutoff frequency: 33.75 Hz)
- Filter length: 497 samples (3.310 sec)
319 events found
Event IDs: [ 1 2 3 4 5 32]
Not setting metadata
Not setting metadata
319 matching events found
Setting baseline interval to [-0.19979521315838786, 0.0] sec
Applying baseline correction (mode: mean)
Created an SSP operator (subspace dimension = 3)
3 projection items activated
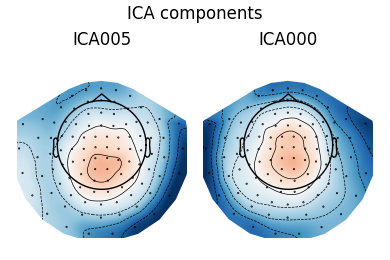
Fit ICA model using the FastICA algorithm, detect and plot components explaining ECG artifacts.
ica = ICA(n_components=0.95, method='fastica').fit(epochs)
ecg_epochs = create_ecg_epochs(raw, tmin=-.5, tmax=.5)
ecg_inds, scores = ica.find_bads_ecg(ecg_epochs, threshold='auto')
ica.plot_components(ecg_inds)
Out:
Fitting ICA to data using 305 channels (please be patient, this may take a while)
Loading data for 319 events and 106 original time points ...
Rejecting epoch based on MAG : ['MEG 1711']
Rejecting epoch based on MAG : ['MEG 1711']
2 bad epochs dropped
Applying projection operator with 3 vectors (pre-whitener computation)
Applying projection operator with 3 vectors (pre-whitener application)
Selecting by explained variance: 126 components
/home/circleci/.local/lib/python3.8/site-packages/sklearn/decomposition/_fastica.py:118: ConvergenceWarning: FastICA did not converge. Consider increasing tolerance or the maximum number of iterations.
warnings.warn('FastICA did not converge. Consider increasing '
Loading data for 317 events and 106 original time points ...
Applying projection operator with 3 vectors (pre-whitener application)
Fitting ICA took 49.3s.
Reconstructing ECG signal from Magnetometers
Setting up band-pass filter from 8 - 16 Hz
FIR filter parameters
---------------------
Designing a two-pass forward and reverse, zero-phase, non-causal bandpass filter:
- Windowed frequency-domain design (firwin2) method
- Hann window
- Lower passband edge: 8.00
- Lower transition bandwidth: 0.50 Hz (-12 dB cutoff frequency: 7.75 Hz)
- Upper passband edge: 16.00 Hz
- Upper transition bandwidth: 0.50 Hz (-12 dB cutoff frequency: 16.25 Hz)
- Filter length: 1502 samples (10.003 sec)
Number of ECG events detected : 284 (average pulse 61 / min.)
Not setting metadata
Not setting metadata
284 matching events found
No baseline correction applied
Created an SSP operator (subspace dimension = 3)
Loading data for 284 events and 151 original time points ...
0 bad epochs dropped
Reconstructing ECG signal from Magnetometers
Using threshold: 0.41 for CTPS ECG detection
Applying projection operator with 3 vectors (pre-whitener application)
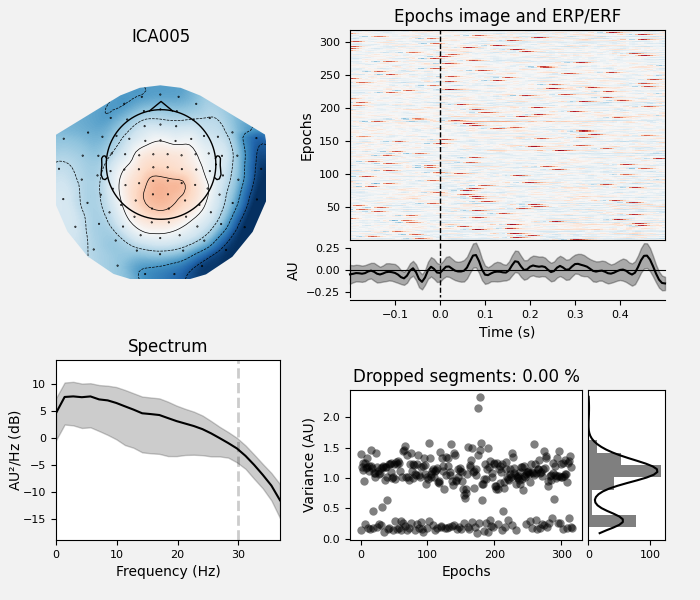
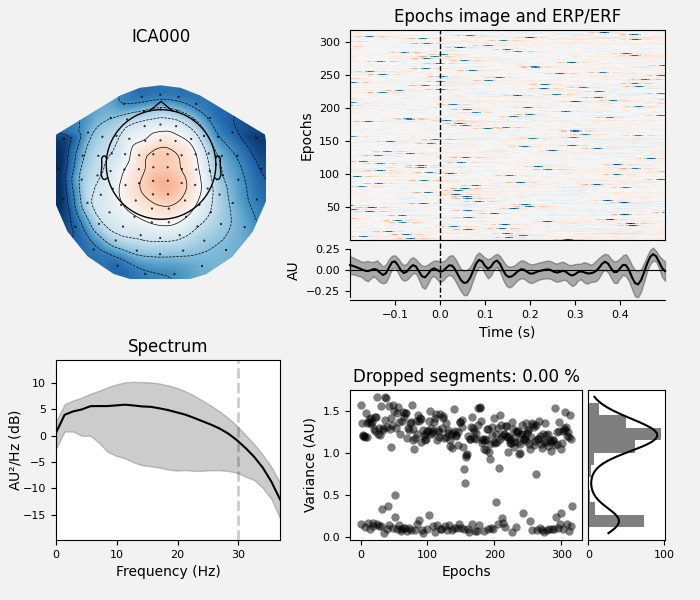
Plot properties of ECG components:
ica.plot_properties(epochs, picks=ecg_inds)
Out:
Loading data for 317 events and 106 original time points ...
Applying projection operator with 3 vectors (pre-whitener application)
Loading data for 317 events and 106 original time points ...
Applying projection operator with 3 vectors (pre-whitener application)
Using multitaper spectrum estimation with 7 DPSS windows
Not setting metadata
Not setting metadata
317 matching events found
No baseline correction applied
0 projection items activated
0 bad epochs dropped
Not setting metadata
Not setting metadata
317 matching events found
No baseline correction applied
0 projection items activated
0 bad epochs dropped
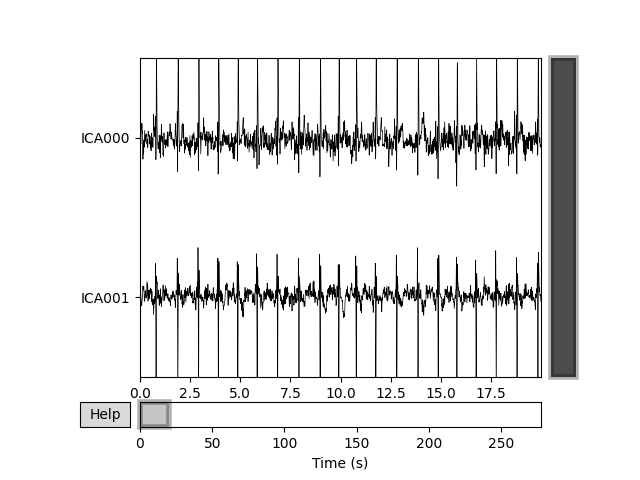
Plot the estimated source of detected ECG related components
ica.plot_sources(raw, picks=ecg_inds)
Out:
Applying projection operator with 3 vectors (pre-whitener application)
Creating RawArray with float64 data, n_channels=2, n_times=41700
Range : 6450 ... 48149 = 42.956 ... 320.665 secs
Ready.
Total running time of the script: ( 1 minutes 5.043 seconds)
Estimated memory usage: 499 MB