Note
Click here to download the full example code
Permutation T-test on sensor data¶
One tests if the signal significantly deviates from 0 during a fixed time window of interest. Here computation is performed on MNE sample dataset between 40 and 60 ms.
# Authors: Alexandre Gramfort <alexandre.gramfort@inria.fr>
#
# License: BSD (3-clause)
import numpy as np
import mne
from mne import io
from mne.stats import permutation_t_test
from mne.datasets import sample
print(__doc__)
Set parameters
data_path = sample.data_path()
raw_fname = data_path + '/MEG/sample/sample_audvis_filt-0-40_raw.fif'
event_fname = data_path + '/MEG/sample/sample_audvis_filt-0-40_raw-eve.fif'
event_id = 1
tmin = -0.2
tmax = 0.5
# Setup for reading the raw data
raw = io.read_raw_fif(raw_fname)
events = mne.read_events(event_fname)
# pick MEG Gradiometers
picks = mne.pick_types(raw.info, meg='grad', eeg=False, stim=False, eog=True,
exclude='bads')
epochs = mne.Epochs(raw, events, event_id, tmin, tmax, picks=picks,
baseline=(None, 0), reject=dict(grad=4000e-13, eog=150e-6))
data = epochs.get_data()
times = epochs.times
temporal_mask = np.logical_and(0.04 <= times, times <= 0.06)
data = np.mean(data[:, :, temporal_mask], axis=2)
n_permutations = 50000
T0, p_values, H0 = permutation_t_test(data, n_permutations, n_jobs=1)
significant_sensors = picks[p_values <= 0.05]
significant_sensors_names = [raw.ch_names[k] for k in significant_sensors]
print("Number of significant sensors : %d" % len(significant_sensors))
print("Sensors names : %s" % significant_sensors_names)
Out:
Opening raw data file /home/circleci/mne_data/MNE-sample-data/MEG/sample/sample_audvis_filt-0-40_raw.fif...
Read a total of 4 projection items:
PCA-v1 (1 x 102) idle
PCA-v2 (1 x 102) idle
PCA-v3 (1 x 102) idle
Average EEG reference (1 x 60) idle
Range : 6450 ... 48149 = 42.956 ... 320.665 secs
Ready.
Not setting metadata
Not setting metadata
72 matching events found
Setting baseline interval to [-0.19979521315838786, 0.0] sec
Applying baseline correction (mode: mean)
4 projection items activated
Loading data for 72 events and 106 original time points ...
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
16 bad epochs dropped
Permuting 49999 times...
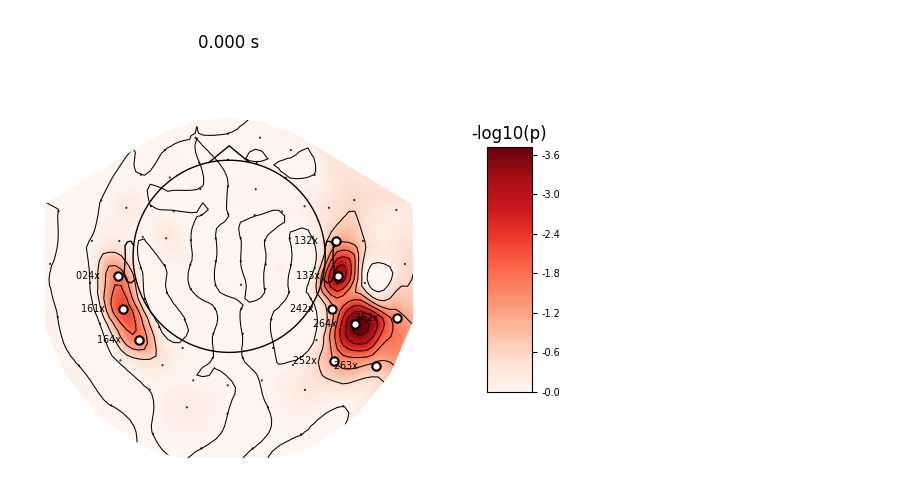
Number of significant sensors : 11
Sensors names : ['MEG 0243', 'MEG 1323', 'MEG 1333', 'MEG 1613', 'MEG 1643', 'MEG 2423', 'MEG 2522', 'MEG 2622', 'MEG 2632', 'MEG 2642', 'MEG 2643']
View location of significantly active sensors
evoked = mne.EvokedArray(-np.log10(p_values)[:, np.newaxis],
epochs.info, tmin=0.)
# Extract mask and indices of active sensors in the layout
stats_picks = mne.pick_channels(evoked.ch_names, significant_sensors_names)
mask = p_values[:, np.newaxis] <= 0.05
evoked.plot_topomap(ch_type='grad', times=[0], scalings=1,
time_format=None, cmap='Reds', vmin=0., vmax=np.max,
units='-log10(p)', cbar_fmt='-%0.1f', mask=mask,
size=3, show_names=lambda x: x[4:] + ' ' * 20,
time_unit='s')
Out:
Removing projector <Projection | PCA-v1, active : True, n_channels : 102>
Removing projector <Projection | PCA-v2, active : True, n_channels : 102>
Removing projector <Projection | PCA-v3, active : True, n_channels : 102>
Removing projector <Projection | Average EEG reference, active : True, n_channels : 60>
Total running time of the script: ( 0 minutes 6.317 seconds)
Estimated memory usage: 185 MB