Note
Click here to download the full example code
Temporal whitening with AR model¶
Here we fit an AR model to the data and use it to temporally whiten the signals.
# Authors: Alexandre Gramfort <alexandre.gramfort@inria.fr>
#
# License: BSD (3-clause)
import numpy as np
from scipy import signal
import matplotlib.pyplot as plt
import mne
from mne.time_frequency import fit_iir_model_raw
from mne.datasets import sample
print(__doc__)
data_path = sample.data_path()
raw_fname = data_path + '/MEG/sample/sample_audvis_raw.fif'
proj_fname = data_path + '/MEG/sample/sample_audvis_ecg-proj.fif'
raw = mne.io.read_raw_fif(raw_fname)
proj = mne.read_proj(proj_fname)
raw.info['projs'] += proj
raw.info['bads'] = ['MEG 2443', 'EEG 053'] # mark bad channels
# Set up pick list: Gradiometers - bad channels
picks = mne.pick_types(raw.info, meg='grad', exclude='bads')
order = 5 # define model order
picks = picks[:1]
# Estimate AR models on raw data
b, a = fit_iir_model_raw(raw, order=order, picks=picks, tmin=60, tmax=180)
d, times = raw[0, 10000:20000] # look at one channel from now on
d = d.ravel() # make flat vector
innovation = signal.convolve(d, a, 'valid')
d_ = signal.lfilter(b, a, innovation) # regenerate the signal
d_ = np.r_[d_[0] * np.ones(order), d_] # dummy samples to keep signal length
Out:
Opening raw data file /home/circleci/mne_data/MNE-sample-data/MEG/sample/sample_audvis_raw.fif...
Read a total of 3 projection items:
PCA-v1 (1 x 102) idle
PCA-v2 (1 x 102) idle
PCA-v3 (1 x 102) idle
Range : 25800 ... 192599 = 42.956 ... 320.670 secs
Ready.
Read a total of 6 projection items:
ECG-planar-999--0.200-0.400-PCA-01 (1 x 203) idle
ECG-planar-999--0.200-0.400-PCA-02 (1 x 203) idle
ECG-axial-999--0.200-0.400-PCA-01 (1 x 102) idle
ECG-axial-999--0.200-0.400-PCA-02 (1 x 102) idle
ECG-eeg-999--0.200-0.400-PCA-01 (1 x 59) idle
ECG-eeg-999--0.200-0.400-PCA-02 (1 x 59) idle
Plot the different time series and PSDs
plt.close('all')
plt.figure()
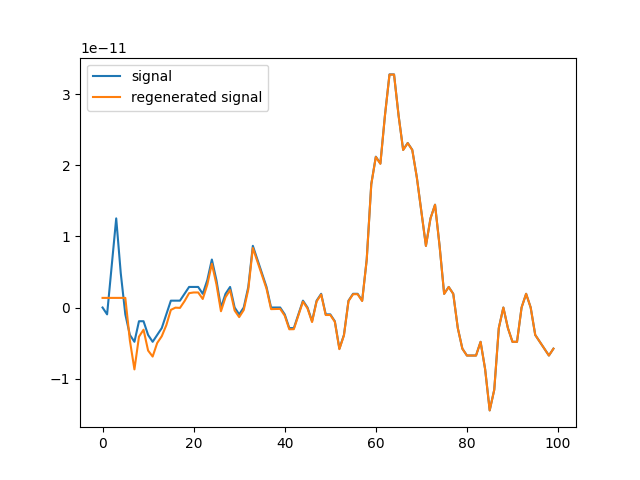
plt.plot(d[:100], label='signal')
plt.plot(d_[:100], label='regenerated signal')
plt.legend()
plt.figure()
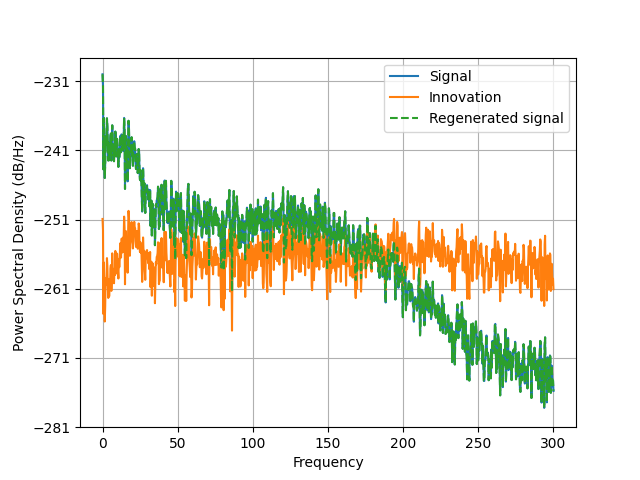
plt.psd(d, Fs=raw.info['sfreq'], NFFT=2048)
plt.psd(innovation, Fs=raw.info['sfreq'], NFFT=2048)
plt.psd(d_, Fs=raw.info['sfreq'], NFFT=2048, linestyle='--')
plt.legend(('Signal', 'Innovation', 'Regenerated signal'))
plt.show()
Total running time of the script: ( 0 minutes 3.656 seconds)
Estimated memory usage: 8 MB