Note
Go to the end to download the full example code.
Convert MNE sample data to BIDS format#
In this example we will use MNE-BIDS to organize the MNE sample data according to the BIDS standard. In a second step we will read the organized dataset using MNE-BIDS.
# Authors: The MNE-BIDS developers
# SPDX-License-Identifier: BSD-3-Clause
First we import some basic Python libraries, followed by MNE-Python and its sample data, and then finally the MNE-BIDS functions we need for this example
import json
import shutil
from pprint import pprint
import mne
from mne.datasets import sample
from mne_bids import (
BIDSPath,
make_dataset_description,
print_dir_tree,
read_raw_bids,
write_meg_calibration,
write_meg_crosstalk,
write_raw_bids,
)
from mne_bids.stats import count_events
Now we can read the MNE sample data. We define an event_id based on our knowledge of the data, to give meaning to events in the data.
With raw_fname and events, we determine where to get the sample data from. output_path determines where we will write the BIDS conversion to.
data_path = sample.data_path()
event_id = {
"Auditory/Left": 1,
"Auditory/Right": 2,
"Visual/Left": 3,
"Visual/Right": 4,
"Smiley": 5,
"Button": 32,
}
raw_fname = data_path / "MEG" / "sample" / "sample_audvis_raw.fif"
er_fname = data_path / "MEG" / "sample" / "ernoise_raw.fif" # empty room
events_fname = data_path / "MEG" / "sample" / "sample_audvis_raw-eve.fif"
output_path = data_path.parent / "MNE-sample-data-bids"
To ensure the output path doesn’t contain any leftover files from previous tests and example runs, we simply delete it.
Warning
Do not delete directories that may contain important data!
Note
mne-bids will try to infer as much information from the data as
possible to then save this data in BIDS-specific “sidecar” files. For
example the manufacturer information, which is inferred from the data file
extension. However, sometimes inferring is ambiguous (e.g., if your file
format is non-standard for the manufacturer). In these cases, MNE-BIDS does
not guess and you will have to update your BIDS fields manually.
Based on our path definitions above, we read the raw data file, define a new BIDS name for it, and then run the automatic BIDS conversion for both the experimental data and its associated empty-room recording.
raw = mne.io.read_raw(raw_fname)
raw_er = mne.io.read_raw(er_fname)
# specify power line frequency as required by BIDS
raw.info["line_freq"] = 60
raw_er.info["line_freq"] = 60
task = "audiovisual"
bids_path = BIDSPath(
subject="01", session="01", task=task, run="1", datatype="meg", root=output_path
)
write_raw_bids(
raw=raw,
bids_path=bids_path,
events=events_fname,
event_id=event_id,
empty_room=raw_er,
overwrite=True,
)
Opening raw data file /home/circleci/mne_data/MNE-sample-data/MEG/sample/sample_audvis_raw.fif...
Read a total of 3 projection items:
PCA-v1 (1 x 102) idle
PCA-v2 (1 x 102) idle
PCA-v3 (1 x 102) idle
Range : 25800 ... 192599 = 42.956 ... 320.670 secs
Ready.
Opening raw data file /home/circleci/mne_data/MNE-sample-data/MEG/sample/ernoise_raw.fif...
Isotrak not found
Read a total of 3 projection items:
PCA-v1 (1 x 102) idle
PCA-v2 (1 x 102) idle
PCA-v3 (1 x 102) idle
Range : 19800 ... 85867 = 32.966 ... 142.965 secs
Ready.
Opening raw data file /home/circleci/mne_data/MNE-sample-data/MEG/sample/sample_audvis_raw.fif...
Read a total of 3 projection items:
PCA-v1 (1 x 102) idle
PCA-v2 (1 x 102) idle
PCA-v3 (1 x 102) idle
Range : 25800 ... 192599 = 42.956 ... 320.670 secs
Ready.
Opening raw data file /home/circleci/mne_data/MNE-sample-data/MEG/sample/ernoise_raw.fif...
Isotrak not found
Read a total of 3 projection items:
PCA-v1 (1 x 102) idle
PCA-v2 (1 x 102) idle
PCA-v3 (1 x 102) idle
Range : 19800 ... 85867 = 32.966 ... 142.965 secs
Ready.
Writing '/home/circleci/mne_data/MNE-sample-data-bids/README'...
Writing '/home/circleci/mne_data/MNE-sample-data-bids/participants.tsv'...
Writing '/home/circleci/mne_data/MNE-sample-data-bids/participants.json'...
Writing of electrodes.tsv is not supported for data type "meg". Skipping ...
Writing '/home/circleci/mne_data/MNE-sample-data-bids/dataset_description.json'...
Writing '/home/circleci/mne_data/MNE-sample-data-bids/sub-emptyroom/ses-20021206/meg/sub-emptyroom_ses-20021206_task-noise_meg.json'...
Copying data files to sub-emptyroom_ses-20021206_task-noise_meg.fif
Writing '/home/circleci/mne_data/MNE-sample-data-bids/sub-emptyroom/ses-20021206/meg/sub-emptyroom_ses-20021206_task-noise_channels.tsv'...
Reserving possible split file sub-emptyroom_ses-20021206_task-noise_split-01_meg.fif
Writing /home/circleci/mne_data/MNE-sample-data-bids/sub-emptyroom/ses-20021206/meg/sub-emptyroom_ses-20021206_task-noise_meg.fif
Closing /home/circleci/mne_data/MNE-sample-data-bids/sub-emptyroom/ses-20021206/meg/sub-emptyroom_ses-20021206_task-noise_meg.fif
[done]
Writing '/home/circleci/mne_data/MNE-sample-data-bids/sub-emptyroom/ses-20021206/sub-emptyroom_ses-20021206_scans.tsv'...
Wrote /home/circleci/mne_data/MNE-sample-data-bids/sub-emptyroom/ses-20021206/sub-emptyroom_ses-20021206_scans.tsv entry with meg/sub-emptyroom_ses-20021206_task-noise_meg.fif.
Writing '/home/circleci/mne_data/MNE-sample-data-bids/participants.tsv'...
Writing '/home/circleci/mne_data/MNE-sample-data-bids/participants.json'...
Writing '/home/circleci/mne_data/MNE-sample-data-bids/sub-01/ses-01/meg/sub-01_ses-01_coordsystem.json'...
Used Annotations descriptions: [np.str_('Auditory/Left'), np.str_('Auditory/Right'), np.str_('Button'), np.str_('Smiley'), np.str_('Visual/Left'), np.str_('Visual/Right')]
Writing '/home/circleci/mne_data/MNE-sample-data-bids/sub-01/ses-01/meg/sub-01_ses-01_task-audiovisual_run-1_events.tsv'...
Writing '/home/circleci/mne_data/MNE-sample-data-bids/sub-01/ses-01/meg/sub-01_ses-01_task-audiovisual_run-1_events.json'...
Writing '/home/circleci/mne_data/MNE-sample-data-bids/dataset_description.json'...
Writing '/home/circleci/mne_data/MNE-sample-data-bids/sub-01/ses-01/meg/sub-01_ses-01_task-audiovisual_run-1_meg.json'...
Copying data files to sub-01_ses-01_task-audiovisual_run-1_meg.fif
Writing '/home/circleci/mne_data/MNE-sample-data-bids/sub-01/ses-01/meg/sub-01_ses-01_task-audiovisual_run-1_channels.tsv'...
Reserving possible split file sub-01_ses-01_task-audiovisual_run-1_split-01_meg.fif
Writing /home/circleci/mne_data/MNE-sample-data-bids/sub-01/ses-01/meg/sub-01_ses-01_task-audiovisual_run-1_meg.fif
Closing /home/circleci/mne_data/MNE-sample-data-bids/sub-01/ses-01/meg/sub-01_ses-01_task-audiovisual_run-1_meg.fif
[done]
Writing '/home/circleci/mne_data/MNE-sample-data-bids/sub-01/ses-01/sub-01_ses-01_scans.tsv'...
Wrote /home/circleci/mne_data/MNE-sample-data-bids/sub-01/ses-01/sub-01_ses-01_scans.tsv entry with meg/sub-01_ses-01_task-audiovisual_run-1_meg.fif.
BIDSPath(
root: /home/circleci/mne_data/MNE-sample-data-bids
datatype: meg
basename: sub-01_ses-01_task-audiovisual_run-1_meg.fif)
Let’s pause and check that the information that we’ve written out to the sidecar files that describe our data is correct.
# Get the sidecar ``.json`` file
sidecar_json_bids_path = bids_path.copy().update(suffix="meg", extension=".json")
sidecar_json_content = sidecar_json_bids_path.fpath.read_text(encoding="utf-8-sig")
print(sidecar_json_content)
{
"TaskName": "audiovisual",
"Manufacturer": "Elekta",
"PowerLineFrequency": 60.0,
"SamplingFrequency": 600.614990234375,
"SoftwareFilters": {
"SpatialCompensation": {
"GradientOrder": 0
}
},
"RecordingDuration": 277.7136813300495,
"RecordingType": "continuous",
"DewarPosition": "n/a",
"DigitizedLandmarks": true,
"DigitizedHeadPoints": true,
"MEGChannelCount": 306,
"MEGREFChannelCount": 0,
"ContinuousHeadLocalization": false,
"HeadCoilFrequency": [],
"AssociatedEmptyRoom": "sub-emptyroom/ses-20021206/meg/sub-emptyroom_ses-20021206_task-noise_meg.fif",
"EEGChannelCount": 60,
"EOGChannelCount": 1,
"ECGChannelCount": 0,
"EMGChannelCount": 0,
"MiscChannelCount": 0,
"TriggerChannelCount": 9
}
The sample MEG dataset comes with fine-calibration and crosstalk files that are required when processing Elekta/Neuromag/MEGIN data using MaxFilter®. Let’s store these data in appropriate places, too.
cal_fname = data_path / "SSS" / "sss_cal_mgh.dat"
ct_fname = data_path / "SSS" / "ct_sparse_mgh.fif"
write_meg_calibration(cal_fname, bids_path)
write_meg_crosstalk(ct_fname, bids_path)
Writing fine-calibration file to /home/circleci/mne_data/MNE-sample-data-bids/sub-01/ses-01/meg/sub-01_ses-01_acq-calibration_meg.dat
Writing crosstalk file to /home/circleci/mne_data/MNE-sample-data-bids/sub-01/ses-01/meg/sub-01_ses-01_acq-crosstalk_meg.fif
Now let’s see the structure of the BIDS folder we created.
|MNE-sample-data-bids/
|--- README
|--- dataset_description.json
|--- participants.json
|--- participants.tsv
|--- sub-01/
|------ ses-01/
|--------- sub-01_ses-01_scans.tsv
|--------- meg/
|------------ sub-01_ses-01_acq-calibration_meg.dat
|------------ sub-01_ses-01_acq-crosstalk_meg.fif
|------------ sub-01_ses-01_coordsystem.json
|------------ sub-01_ses-01_task-audiovisual_run-1_channels.tsv
|------------ sub-01_ses-01_task-audiovisual_run-1_events.json
|------------ sub-01_ses-01_task-audiovisual_run-1_events.tsv
|------------ sub-01_ses-01_task-audiovisual_run-1_meg.fif
|------------ sub-01_ses-01_task-audiovisual_run-1_meg.json
|--- sub-emptyroom/
|------ ses-20021206/
|--------- sub-emptyroom_ses-20021206_scans.tsv
|--------- meg/
|------------ sub-emptyroom_ses-20021206_task-noise_channels.tsv
|------------ sub-emptyroom_ses-20021206_task-noise_meg.fif
|------------ sub-emptyroom_ses-20021206_task-noise_meg.json
Now let’s get an overview of the events on the whole dataset
A big advantage of having data organized according to BIDS is that software
packages can automate your workflow. For example, reading the data back
into MNE-Python can easily be done using read_raw_bids().
Opening raw data file /home/circleci/mne_data/MNE-sample-data-bids/sub-01/ses-01/meg/sub-01_ses-01_task-audiovisual_run-1_meg.fif...
Read a total of 3 projection items:
PCA-v1 (1 x 102) idle
PCA-v2 (1 x 102) idle
PCA-v3 (1 x 102) idle
Range : 25800 ... 192599 = 42.956 ... 320.670 secs
Ready.
Reading events from /home/circleci/mne_data/MNE-sample-data-bids/sub-01/ses-01/meg/sub-01_ses-01_task-audiovisual_run-1_events.tsv.
Reading channel info from /home/circleci/mne_data/MNE-sample-data-bids/sub-01/ses-01/meg/sub-01_ses-01_task-audiovisual_run-1_channels.tsv.
The resulting data is already in a convenient form to create epochs and evoked data.
events, event_id = mne.events_from_annotations(raw)
epochs = mne.Epochs(raw, events, event_id)
epochs["Auditory"].average().plot()
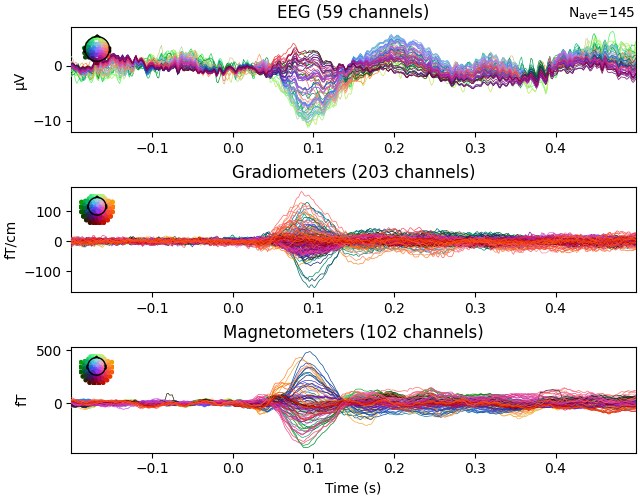
Used Annotations descriptions: [np.str_('Auditory/Left'), np.str_('Auditory/Right'), np.str_('Button'), np.str_('Smiley'), np.str_('Visual/Left'), np.str_('Visual/Right')]
Not setting metadata
320 matching events found
Setting baseline interval to [-0.19979521315838786, 0.0] s
Applying baseline correction (mode: mean)
Created an SSP operator (subspace dimension = 3)
3 projection items activated
<Figure size 640x500 with 6 Axes>
We can easily get the mne_bids.BIDSPath of the empty-room recording
that was associated with the experimental data while writing. The empty-room
data can then be loaded with read_raw_bids().
er_bids_path = bids_path.find_empty_room(use_sidecar_only=True)
er_data = read_raw_bids(er_bids_path)
er_data
Using "AssociatedEmptyRoom" entry from MEG sidecar file to retrieve empty-room path.
Opening raw data file /home/circleci/mne_data/MNE-sample-data-bids/sub-emptyroom/ses-20021206/meg/sub-emptyroom_ses-20021206_task-noise_meg.fif...
Isotrak not found
Read a total of 3 projection items:
PCA-v1 (1 x 102) idle
PCA-v2 (1 x 102) idle
PCA-v3 (1 x 102) idle
Range : 19800 ... 85867 = 32.966 ... 142.965 secs
Ready.
Reading channel info from /home/circleci/mne_data/MNE-sample-data-bids/sub-emptyroom/ses-20021206/meg/sub-emptyroom_ses-20021206_task-noise_channels.tsv.
It is trivial to retrieve the path of the fine-calibration and crosstalk files, too.
print(bids_path.meg_calibration_fpath)
print(bids_path.meg_crosstalk_fpath)
/home/circleci/mne_data/MNE-sample-data-bids/sub-01/ses-01/meg/sub-01_ses-01_acq-calibration_meg.dat
/home/circleci/mne_data/MNE-sample-data-bids/sub-01/ses-01/meg/sub-01_ses-01_acq-crosstalk_meg.fif
The README created by write_raw_bids() also takes care of the citation
for mne-bids. If you are preparing a manuscript, please make sure to also
cite MNE-BIDS there.
readme = output_path / "README"
with open(readme, encoding="utf-8-sig") as fid:
text = fid.read()
print(text)
References
----------
Appelhoff, S., Sanderson, M., Brooks, T., Vliet, M., Quentin, R., Holdgraf, C., Chaumon, M., Mikulan, E., Tavabi, K., Höchenberger, R., Welke, D., Brunner, C., Rockhill, A., Larson, E., Gramfort, A. and Jas, M. (2019). MNE-BIDS: Organizing electrophysiological data into the BIDS format and facilitating their analysis. Journal of Open Source Software 4: (1896).https://doi.org/10.21105/joss.01896
Niso, G., Gorgolewski, K. J., Bock, E., Brooks, T. L., Flandin, G., Gramfort, A., Henson, R. N., Jas, M., Litvak, V., Moreau, J., Oostenveld, R., Schoffelen, J., Tadel, F., Wexler, J., Baillet, S. (2018). MEG-BIDS, the brain imaging data structure extended to magnetoencephalography. Scientific Data, 5, 180110.https://doi.org/10.1038/sdata.2018.110
It is also generally a good idea to add a description of your dataset, see the BIDS dataset_description.json definition for more information.
how_to_acknowledge = """\
If you reference this dataset in a publication, please acknowledge its \
authors and cite MNE papers: A. Gramfort, M. Luessi, E. Larson, D. Engemann, \
D. Strohmeier, C. Brodbeck, L. Parkkonen, M. Hämäläinen, \
MNE software for processing MEG and EEG data, NeuroImage, Volume 86, \
1 February 2014, Pages 446-460, ISSN 1053-8119 \
and \
A. Gramfort, M. Luessi, E. Larson, D. Engemann, D. Strohmeier, C. Brodbeck, \
R. Goj, M. Jas, T. Brooks, L. Parkkonen, M. Hämäläinen, MEG and EEG data \
analysis with MNE-Python, Frontiers in Neuroscience, Volume 7, 2013, \
ISSN 1662-453X"""
make_dataset_description(
path=bids_path.root,
name=task,
authors=["Alexandre Gramfort", "Matti Hämäläinen"],
how_to_acknowledge=how_to_acknowledge,
acknowledgements="""\
Alexandre Gramfort, Mainak Jas, and Stefan Appelhoff prepared and updated the \
data in BIDS format.""",
data_license="CC0",
ethics_approvals=["Human Subjects Division at the University of Washington"],
funding=[
"NIH 5R01EB009048",
"NIH 1R01EB009048",
"NIH R01EB006385",
"NIH 1R01HD40712",
"NIH 1R01NS44319",
"NIH 2R01NS37462",
"NIH P41EB015896",
"ANR-11-IDEX-0003-02",
"ERC-StG-263584",
"ERC-StG-676943",
"ANR-14-NEUC-0002-01",
],
references_and_links=[
"https://doi.org/10.1016/j.neuroimage.2014.02.017",
"https://doi.org/10.3389/fnins.2013.00267",
"https://mne.tools/stable/documentation/datasets.html#sample",
],
doi="doi:10.18112/openneuro.ds000248.v1.2.4",
overwrite=True,
)
desc_json_path = bids_path.root / "dataset_description.json"
with open(desc_json_path, encoding="utf-8-sig") as fid:
pprint(json.loads(fid.read()))
Writing '/home/circleci/mne_data/MNE-sample-data-bids/dataset_description.json'...
{'Acknowledgements': 'Alexandre Gramfort, Mainak Jas, and Stefan Appelhoff '
'prepared and updated the data in BIDS format.',
'Authors': ['Alexandre Gramfort', 'Matti Hämäläinen'],
'BIDSVersion': '1.9.0',
'DatasetDOI': 'doi:10.18112/openneuro.ds000248.v1.2.4',
'DatasetType': 'raw',
'EthicsApprovals': ['Human Subjects Division at the University of Washington'],
'Funding': ['NIH 5R01EB009048',
'NIH 1R01EB009048',
'NIH R01EB006385',
'NIH 1R01HD40712',
'NIH 1R01NS44319',
'NIH 2R01NS37462',
'NIH P41EB015896',
'ANR-11-IDEX-0003-02',
'ERC-StG-263584',
'ERC-StG-676943',
'ANR-14-NEUC-0002-01'],
'GeneratedBy': [{'CodeURL': 'https://mne.tools/mne-bids/',
'Name': 'MNE-BIDS',
'Version': '0.19.0.dev20+ge66a9ea7a'}],
'HowToAcknowledge': 'If you reference this dataset in a publication, please '
'acknowledge its authors and cite MNE papers: A. '
'Gramfort, M. Luessi, E. Larson, D. Engemann, D. '
'Strohmeier, C. Brodbeck, L. Parkkonen, M. Hämäläinen, '
'MNE software for processing MEG and EEG data, '
'NeuroImage, Volume 86, 1 February 2014, Pages 446-460, '
'ISSN 1053-8119 and A. Gramfort, M. Luessi, E. Larson, D. '
'Engemann, D. Strohmeier, C. Brodbeck, R. Goj, M. Jas, T. '
'Brooks, L. Parkkonen, M. Hämäläinen, MEG and EEG data '
'analysis with MNE-Python, Frontiers in Neuroscience, '
'Volume 7, 2013, ISSN 1662-453X',
'License': 'CC0',
'Name': 'audiovisual',
'ReferencesAndLinks': ['https://doi.org/10.1016/j.neuroimage.2014.02.017',
'https://doi.org/10.3389/fnins.2013.00267',
'https://mne.tools/stable/documentation/datasets.html#sample']}
This should be very similar to the ds000248 dataset_description.json!
Total running time of the script: (0 minutes 3.597 seconds)