Note
Click here to download the full example code or to run this example in your browser via Binder
08. Convert iEEG data to BIDS format¶
In this example, we use MNE-BIDS to create a BIDS-compatible directory of iEEG data. Specifically, we will follow these steps:
Download some iEEG data from the MNE-ECoG example.
Load the data, extract information, and save in a new BIDS directory.
Check the result and compare it with the standard.
Cite MNE-BIDS
Confirm that written iEEG coordinates are the same before
write_raw_bids()was called.
The iEEG data will be written by write_raw_bids() with
the addition of extra metadata elements in the following files:
the sidecar file
ieeg.jsonelectrodes.tsvcoordsystem.jsonevents.tsvchannels.tsv
Compared to EEG data, the main differences are within the
coordsystem.json and electrodes.tsv files.
For more information on these files,
refer to the iEEG part of the BIDS specification.
# Authors: Adam Li <adam2392@gmail.com>
# Stefan Appelhoff <stefan.appelhoff@mailbox.org>
#
# License: BSD (3-clause)
import os.path as op
import shutil
from pprint import pprint
import numpy as np
import mne
from mne_bids import (write_raw_bids, BIDSPath,
read_raw_bids, print_dir_tree)
Step 1: Download the data¶
First, we need some data to work with. We will use the
data downloaded via MNE-Python’s datasets API:
mne.datasets.misc.data_path()
misc_path = mne.datasets.misc.data_path(force_update=True)
# The electrode coords data are in the tsv file format
# which is easily read in using numpy
fname = misc_path + '/ecog/sample_ecog_electrodes.tsv'
data = np.loadtxt(fname, dtype=str, delimiter='\t',
comments=None, encoding='utf-8')
column_names = data[0, :]
info = data[1:, :]
electrode_tsv = dict()
for i, name in enumerate(column_names):
electrode_tsv[name] = info[:, i].tolist()
# load in channel names
ch_names = electrode_tsv['name']
# load in the xyz coordinates as a float
elec = np.empty(shape=(len(ch_names), 3))
for ind, axis in enumerate(['x', 'y', 'z']):
elec[:, ind] = list(map(float, electrode_tsv[axis]))
Out:
Downloading archive mne-misc-data-0.8.tar.gz to /Users/hoechenberger/mne_data
Downloading https://codeload.github.com/mne-tools/mne-misc-data/tar.gz/0.8 (0 bytes)
0%| | Downloading : 0.00/0.00 [00:00<?, ?B/s]
0%| | Downloading : 120k/0.00 [00:00<00:00, 5.41MB/s]
0%| | Downloading : 312k/0.00 [00:00<00:00, 8.00MB/s]
0%| | Downloading : 568k/0.00 [00:00<00:00, 10.1MB/s]
0%| | Downloading : 1.30M/0.00 [00:00<00:00, 16.2MB/s]
0%| | Downloading : 1.80M/0.00 [00:00<00:00, 18.9MB/s]
0%| | Downloading : 2.30M/0.00 [00:00<00:00, 19.2MB/s]
0%| | Downloading : 2.80M/0.00 [00:00<00:00, 19.4MB/s]
0%| | Downloading : 3.30M/0.00 [00:00<00:00, 9.59MB/s]
0%| | Downloading : 3.55M/0.00 [00:00<00:00, 8.11MB/s]
0%| | Downloading : 3.80M/0.00 [00:00<00:00, 6.78MB/s]
0%| | Downloading : 3.93M/0.00 [00:00<00:00, 6.19MB/s]
0%| | Downloading : 4.05M/0.00 [00:00<00:00, 5.88MB/s]
0%| | Downloading : 4.18M/0.00 [00:00<00:00, 5.63MB/s]
0%| | Downloading : 4.30M/0.00 [00:00<00:00, 5.57MB/s]
0%| | Downloading : 4.68M/0.00 [00:00<00:00, 6.05MB/s]
0%| | Downloading : 5.18M/0.00 [00:00<00:00, 6.71MB/s]
0%| | Downloading : 5.68M/0.00 [00:00<00:00, 7.20MB/s]
0%| | Downloading : 6.18M/0.00 [00:00<00:00, 7.82MB/s]
0%| | Downloading : 6.68M/0.00 [00:00<00:00, 8.38MB/s]
0%| | Downloading : 6.93M/0.00 [00:00<00:00, 8.50MB/s]
0%| | Downloading : 7.43M/0.00 [00:00<00:00, 9.07MB/s]
0%| | Downloading : 7.93M/0.00 [00:00<00:00, 9.58MB/s]
0%| | Downloading : 8.43M/0.00 [00:00<00:00, 9.90MB/s]
0%| | Downloading : 8.93M/0.00 [00:01<00:00, 10.4MB/s]
0%| | Downloading : 9.43M/0.00 [00:01<00:00, 10.8MB/s]
0%| | Downloading : 9.68M/0.00 [00:01<00:00, 10.9MB/s]
0%| | Downloading : 10.2M/0.00 [00:01<00:00, 11.3MB/s]
0%| | Downloading : 10.7M/0.00 [00:01<00:00, 11.6MB/s]
0%| | Downloading : 11.2M/0.00 [00:01<00:00, 11.9MB/s]
0%| | Downloading : 11.7M/0.00 [00:01<00:00, 12.3MB/s]
0%| | Downloading : 12.2M/0.00 [00:01<00:00, 12.6MB/s]
0%| | Downloading : 12.4M/0.00 [00:01<00:00, 12.6MB/s]
0%| | Downloading : 12.9M/0.00 [00:01<00:00, 13.0MB/s]
0%| | Downloading : 13.4M/0.00 [00:01<00:00, 11.9MB/s]
0%| | Downloading : 13.7M/0.00 [00:01<00:00, 10.4MB/s]
0%| | Downloading : 13.8M/0.00 [00:01<00:00, 9.73MB/s]
0%| | Downloading : 13.9M/0.00 [00:01<00:00, 8.94MB/s]
0%| | Downloading : 14.1M/0.00 [00:01<00:00, 8.62MB/s]
0%| | Downloading : 14.2M/0.00 [00:01<00:00, 8.33MB/s]
0%| | Downloading : 14.3M/0.00 [00:01<00:00, 7.87MB/s]
0%| | Downloading : 14.4M/0.00 [00:01<00:00, 7.63MB/s]
0%| | Downloading : 14.6M/0.00 [00:01<00:00, 7.39MB/s]
0%| | Downloading : 14.9M/0.00 [00:01<00:00, 7.69MB/s]
0%| | Downloading : 15.4M/0.00 [00:01<00:00, 8.12MB/s]
0%| | Downloading : 15.9M/0.00 [00:01<00:00, 8.41MB/s]
0%| | Downloading : 16.4M/0.00 [00:01<00:00, 8.86MB/s]
0%| | Downloading : 16.9M/0.00 [00:01<00:00, 9.25MB/s]
0%| | Downloading : 17.2M/0.00 [00:01<00:00, 9.35MB/s]
0%| | Downloading : 17.4M/0.00 [00:01<00:00, 9.49MB/s]
0%| | Downloading : 18.2M/0.00 [00:02<00:00, 10.1MB/s]
0%| | Downloading : 18.7M/0.00 [00:02<00:00, 10.4MB/s]
0%| | Downloading : 19.2M/0.00 [00:02<00:00, 10.7MB/s]
0%| | Downloading : 19.7M/0.00 [00:02<00:00, 11.1MB/s]
0%| | Downloading : 20.2M/0.00 [00:02<00:00, 11.3MB/s]
0%| | Downloading : 20.7M/0.00 [00:02<00:00, 11.6MB/s]
0%| | Downloading : 21.2M/0.00 [00:02<00:00, 11.9MB/s]
0%| | Downloading : 21.7M/0.00 [00:02<00:00, 12.1MB/s]
0%| | Downloading : 22.2M/0.00 [00:02<00:00, 12.3MB/s]
0%| | Downloading : 22.7M/0.00 [00:02<00:00, 12.5MB/s]
0%| | Downloading : 23.2M/0.00 [00:02<00:00, 12.8MB/s]
0%| | Downloading : 23.7M/0.00 [00:02<00:00, 13.1MB/s]
0%| | Downloading : 24.2M/0.00 [00:02<00:00, 13.2MB/s]
0%| | Downloading : 24.7M/0.00 [00:02<00:00, 13.5MB/s]
0%| | Downloading : 25.2M/0.00 [00:02<00:00, 13.8MB/s]
0%| | Downloading : 25.7M/0.00 [00:02<00:00, 13.8MB/s]
0%| | Downloading : 26.2M/0.00 [00:02<00:00, 14.1MB/s]
0%| | Downloading : 26.7M/0.00 [00:02<00:00, 14.1MB/s]
0%| | Downloading : 27.2M/0.00 [00:02<00:00, 14.2MB/s]
0%| | Downloading : 27.7M/0.00 [00:02<00:00, 14.5MB/s]
0%| | Downloading : 28.2M/0.00 [00:02<00:00, 14.6MB/s]
0%| | Downloading : 28.7M/0.00 [00:02<00:00, 14.8MB/s]
0%| | Downloading : 29.2M/0.00 [00:02<00:00, 15.1MB/s]
0%| | Downloading : 29.7M/0.00 [00:02<00:00, 15.0MB/s]
0%| | Downloading : 30.2M/0.00 [00:02<00:00, 15.2MB/s]
0%| | Downloading : 30.7M/0.00 [00:02<00:00, 15.2MB/s]
0%| | Downloading : 30.9M/0.00 [00:02<00:00, 15.0MB/s]
0%| | Downloading : 30.9M/0.00 [00:02<00:00, 11.7MB/s]
Verifying hash 0f88194266121dd9409be94184231f25.
Removing old directory: /Users/hoechenberger/mne_data/MNE-misc-data
Decompressing the archive: /Users/hoechenberger/mne_data/mne-misc-data-0.8.tar.gz
(please be patient, this can take some time)
Successfully extracted to: ['/Users/hoechenberger/mne_data/MNE-misc-data']
Now we make a montage stating that the iEEG contacts are in the MNI coordinate system, which corresponds to the fsaverage subject in FreeSurfer. For example, one can look at how MNE-Python deals with iEEG data at Working with SEEG.
Out:
Created 64 channel positions
{'C1': array([-0.0132, -0.0654, 0.0702]), 'C2': array([-0.0145, -0.0594, 0.0781]), 'C3': array([-0.0156, -0.0518, 0.0838]), 'C4': array([-0.0154, -0.0453, 0.0895]), 'C5': array([-0.0149, -0.0385, 0.094 ]), 'C6': array([-0.0131, -0.029 , 0.0985]), 'C7': array([-0.0136, -0.0203, 0.1009]), 'C8': array([-0.0109, -0.0093, 0.1035]), 'C9': array([-0.0215, -0.0664, 0.0691]), 'C10': array([-0.0233, -0.0607, 0.0769]), 'C11': array([-0.0247, -0.0534, 0.0822]), 'C12': array([-0.0263, -0.0461, 0.0871]), 'C13': array([-0.0261, -0.0383, 0.0918]), 'C14': array([-0.0253, -0.0299, 0.0949]), 'C15': array([-0.0233, -0.0193, 0.0989]), 'C16': array([-0.0211, -0.008 , 0.1026]), 'C17': array([-0.0304, -0.0674, 0.0678]), 'C18': array([-0.0328, -0.061 , 0.0738]), 'C19': array([-0.0337, -0.0539, 0.0793]), 'C20': array([-0.0344, -0.0466, 0.0842]), 'C21': array([-0.0363, -0.0388, 0.0878]), 'C22': array([-0.0345, -0.0288, 0.0926]), 'C23': array([-0.0335, -0.018 , 0.0955]), 'C24': array([-0.0299, -0.008 , 0.0977]), 'C25': array([-0.0395, -0.0661, 0.0632]), 'C26': array([-0.0417, -0.0599, 0.069 ]), 'C27': array([-0.0426, -0.0529, 0.0744]), 'C28': array([-0.0438, -0.0456, 0.0795]), 'C29': array([-0.0457, -0.0376, 0.083 ]), 'C30': array([-0.0446, -0.0273, 0.0881]), 'C31': array([-0.043 , -0.0176, 0.0901]), 'C32': array([-0.0396, -0.0063, 0.0884]), 'C33': array([-0.0446, -0.0657, 0.0581]), 'C34': array([-0.0482, -0.0593, 0.0632]), 'C35': array([-0.0505, -0.0523, 0.0694]), 'C36': array([-0.0536, -0.0438, 0.0731]), 'C37': array([-0.0548, -0.0357, 0.0769]), 'C38': array([-0.0533, -0.0254, 0.0808]), 'C39': array([-0.0512, -0.0156, 0.0823]), 'C40': array([-0.0473, -0.0044, 0.0808]), 'C41': array([-0.0518, -0.0637, 0.0513]), 'C42': array([-0.055 , -0.0572, 0.0574]), 'C43': array([-0.0575, -0.0501, 0.0625]), 'C44': array([-0.0606, -0.0414, 0.0666]), 'C45': array([-0.0614, -0.0334, 0.0709]), 'C46': array([-0.0612, -0.023 , 0.0733]), 'C47': array([-0.0589, -0.0135, 0.0743]), 'C48': array([-0.0553, -0.0025, 0.0739]), 'C49': array([-0.0575, -0.0605, 0.0455]), 'C50': array([-0.0615, -0.0536, 0.05 ]), 'C51': array([-0.0635, -0.0474, 0.0554]), 'C52': array([-0.066 , -0.0385, 0.0584]), 'C53': array([-0.0671, -0.0306, 0.063 ]), 'C54': array([-0.0672, -0.0204, 0.0644]), 'C55': array([-0.0643, -0.0112, 0.0638]), 'C56': array([-0.0602, -0.0007, 0.064 ]), 'C57': array([-0.0614, -0.0574, 0.0372]), 'C58': array([-0.0659, -0.05 , 0.0412]), 'C59': array([-0.0681, -0.0434, 0.0463]), 'C60': array([-0.0694, -0.0357, 0.0503]), 'C61': array([-0.0704, -0.0277, 0.053 ]), 'C62': array([-0.0705, -0.0177, 0.0541]), 'C63': array([-0.068 , -0.0074, 0.053 ]), 'C64': array([-0.0642, 0.0016, 0.0539])}
We will load a mne.io.Raw object and
use the montage we created.
info = mne.create_info(ch_names, 1000., 'ecog')
raw = mne.io.read_raw_edf(misc_path + '/ecog/sample_ecog.edf')
raw.info['line_freq'] = 60 # specify power line frequency as required by BIDS
raw.set_channel_types({ch: 'ecog' for ch in raw.ch_names})
# set the bad channels
raw.info['bads'].extend(['BTM1', 'BTM2', 'BTM3', 'BTM4', 'BTM5', 'BTM6',
'BTP1', 'BTP2', 'BTP3', 'BTP4', 'BTP5', 'BTP6',
'EKGL', 'EKGR'])
# set montage
raw.set_montage(montage, on_missing='warn')
Out:
Extracting EDF parameters from /Users/hoechenberger/mne_data/MNE-misc-data/ecog/sample_ecog.edf...
EDF file detected
Setting channel info structure...
Creating raw.info structure...
/Users/hoechenberger/Development/mne-bids/examples/convert_ieeg_to_bids.py:113: RuntimeWarning: Fiducial point nasion not found, assuming identity unknown to head transformation
raw.set_montage(montage, on_missing='warn')
/Users/hoechenberger/Development/mne-bids/examples/convert_ieeg_to_bids.py:113: RuntimeWarning: DigMontage is only a subset of info. There are 14 channel positions not present in the DigMontage. The required channels are:
['BTM1', 'BTM2', 'BTM3', 'BTM4', 'BTM5', 'BTM6', 'BTP1', 'BTP2', 'BTP3', 'BTP4', 'BTP5', 'BTP6', 'EKGL', 'EKGR'].
Consider using inst.set_channel_types if these are not EEG channels, or use the on_missing parameter if the channel positions are allowed to be unknown in your analyses.
raw.set_montage(montage, on_missing='warn')
Let us confirm what our channel coordinates look like.
# make a plot of the sensors in 2D plane
raw.plot_sensors(ch_type='ecog')
# Get the first 5 channels and show their locations.
picks = mne.pick_types(raw.info, ecog=True)
dig = [raw.info['dig'][pick] for pick in picks]
chs = [raw.info['chs'][pick] for pick in picks]
pos = np.array([ch['r'] for ch in dig[:5]])
ch_names = np.array([ch['ch_name'] for ch in chs[:5]])
print("The channel coordinates before writing into BIDS: ")
pprint([x for x in zip(ch_names, pos)])
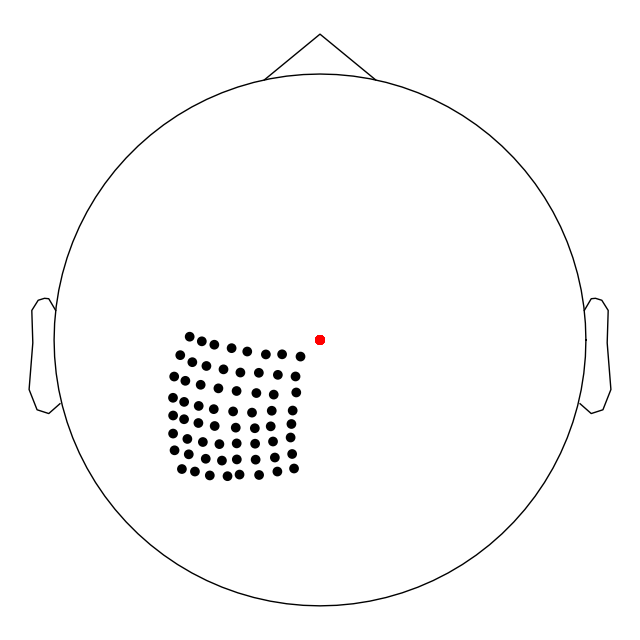
Out:
The channel coordinates before writing into BIDS:
[('C1', array([-0.0132, -0.0654, 0.0702])),
('C2', array([-0.0145, -0.0594, 0.0781])),
('C3', array([-0.0156, -0.0518, 0.0838])),
('C4', array([-0.0154, -0.0453, 0.0895])),
('C5', array([-0.0149, -0.0385, 0.094 ]))]
BIDS vs MNE-Python Coordinate Systems¶
BIDS has many acceptable coordinate systems for iEEG, which can be viewed in appendix VIII of the BIDS specification. However, MNE-BIDS depends on MNE-Python and MNE-Python does not support all these coordinate systems (yet).
MNE-Python has a few tutorials on this topic:
Currently, MNE-Python supports the mni_tal coordinate frame, which
corresponds to the fsaverage BIDS coordinate system. All other coordinate
frames in MNE-Python if written with mne_bids.write_raw_bids() are
written with coordinate system 'Other'. Note, then we suggest using
mne_bids.update_sidecar_json() to update the sidecar
*_coordsystem.json file to add additional information.
Step 2: Formatting as BIDS¶
Now, let us format the Raw object into BIDS.
With this step, we have everything to start a new BIDS directory using
our data. To do that, we can use write_raw_bids()
Generally, write_raw_bids() tries to extract as much
meta data as possible from the raw data and then formats it in a BIDS
compatible way. write_raw_bids() takes a bunch of inputs, most of
which are however optional. The required inputs are:
rawbids_basenamebids_root
… as you can see in the docstring:
print(write_raw_bids.__doc__)
Out:
Save raw data to a BIDS-compliant folder structure.
.. warning:: * The original file is simply copied over if the original
file format is BIDS-supported for that datatype. Otherwise,
this function will convert to a BIDS-supported file format
while warning the user. For EEG and iEEG data, conversion
will be to BrainVision format; for MEG, conversion will be
to FIFF.
* ``mne-bids`` will infer the manufacturer information
from the file extension. If your file format is non-standard
for the manufacturer, please update the manufacturer field
in the sidecars manually.
Parameters
----------
raw : mne.io.Raw
The raw data. It must be an instance of `mne.io.Raw`. The data
should not be loaded from disk, i.e., ``raw.preload`` must be
``False``.
bids_path : mne_bids.BIDSPath
The file to write. The `mne_bids.BIDSPath` instance passed here
**must** have the ``.root`` attribute set. If the ``.datatype``
attribute is not set, it will be inferred from the recording data type
found in ``raw``.
Example::
bids_path = BIDSPath(subject='01', session='01', task='testing',
acquisition='01', run='01', datatype='meg',
root='/data/BIDS')
This will write the following files in the correct subfolder ``root``::
sub-01_ses-01_task-testing_acq-01_run-01_meg.fif
sub-01_ses-01_task-testing_acq-01_run-01_meg.json
sub-01_ses-01_task-testing_acq-01_run-01_channels.tsv
sub-01_ses-01_acq-01_coordsystem.json
and the following one if ``events_data`` is not ``None``::
sub-01_ses-01_task-testing_acq-01_run-01_events.tsv
and add a line to the following files::
participants.tsv
scans.tsv
Note that the data type is automatically inferred from the raw
object, as well as the extension. Data with MEG and other
electrophysiology data in the same file will be stored as ``'meg'``.
events_data : path-like | np.ndarray | None
Use this parameter to specify events to write to the ``*_events.tsv``
sidecar file, additionally to the object's `mne.Annotations` (which
are always written).
If a path, specifies the location of an MNE events file.
If an array, the MNE events array (shape: ``(n_events, 3)``).
If a path or an array and ``raw.annotations`` exist, the union of
``event_data`` and ``raw.annotations`` will be written.
Corresponding descriptions for all event IDs (listed in the third
column of the MNE events array) must be specified via the ``event_id``
parameter; otherwise, an exception is raised.
If ``None``, events will only be inferred from the the raw object's
`mne.Annotations`.
.. note::
If ``not None``, writes the union of ``events_data`` and
``raw.annotations``. If you wish to **only** write
``raw.annotations``, pass ``events_data=None``. If you want to
**exclude** the events in ``raw.annotations`` from being written,
call ``raw.set_annotations(None)`` before invoking this function.
.. note::
Descriptions of all event IDs must be specified via the ``event_id``
parameter.
event_id : dict | None
Descriptions of all event IDs, if you passed ``events_data``.
The descriptions will be written to the ``trial_type`` column in
``*_events.tsv``. The dictionary keys correspond to the event
descriptions and the values to the event IDs. You must specify a
description for all event IDs in ``events_data``.
anonymize : dict | None
If `None` (default), no anonymization is performed.
If a dictionary, data will be anonymized depending on the dictionary
keys: ``daysback`` is a required key, ``keep_his`` is optional.
``daysback`` : int
Number of days by which to move back the recording date in time.
In studies with multiple subjects the relative recording date
differences between subjects can be kept by using the same number
of ``daysback`` for all subject anonymizations. ``daysback`` should
be great enough to shift the date prior to 1925 to conform with
BIDS anonymization rules.
``keep_his`` : bool
If ``False`` (default), all subject information next to the
recording date will be overwritten as well. If True, keep subject
information apart from the recording date.
format : 'auto' | 'BrainVision' | 'FIF'
Controls the file format of the data after BIDS conversion. If
``'auto'``, MNE-BIDS will attempt to convert the input data to BIDS
without a change of the original file format. A conversion to a
different file format (BrainVision, or FIF) will only take place when
the original file format lacks some necessary features. When a str is
passed, a conversion can be forced to the BrainVision format for EEG,
or the FIF format for MEG data.
overwrite : bool
Whether to overwrite existing files or data in files.
Defaults to ``False``.
If ``True``, any existing files with the same BIDS parameters
will be overwritten with the exception of the ``*_participants.tsv``
and ``*_scans.tsv`` files. For these files, parts of pre-existing data
that match the current data will be replaced. For
``*_participants.tsv``, specifically, age, sex and hand fields will be
overwritten, while any manually added fields in ``participants.json``
and ``participants.tsv`` by a user will be retained.
If ``False``, no existing data will be overwritten or
replaced.
verbose : bool
If ``True``, this will print a snippet of the sidecar files. Otherwise,
no content will be printed.
Returns
-------
bids_path : mne_bids.BIDSPath
The path of the created data file.
Notes
-----
You should ensure that ``raw.info['subject_info']`` and
``raw.info['meas_date']`` are set to proper (not-``None``) values to allow
for the correct computation of each participant's age when creating
``*_participants.tsv``.
This function will convert existing `mne.Annotations` from
``raw.annotations`` to events. Additionally, any events supplied via
``events_data`` will be written too. To avoid writing of annotations,
remove them from the raw file via ``raw.set_annotations(None)`` before
invoking ``write_raw_bids``.
To write events encoded in a ``STIM`` channel, you first need to create the
events array manually and pass it to this function:
..
events = mne.find_events(raw, min_duration=0.002)
write_raw_bids(..., events_data=events)
See the documentation of `mne.find_events` for more information on event
extraction from ``STIM`` channels.
When anonymizing ``.edf`` files, then the file format for EDF limits
how far back we can set the recording date. Therefore, all anonymized
EDF datasets will have an internal recording date of ``01-01-1985``,
and the actual recording date will be stored in the ``scans.tsv``
file's ``acq_time`` column.
See Also
--------
mne.io.Raw.anonymize
mne.find_events
mne.Annotations
mne.events_from_annotations
Let us initialize some of the necessary data for the subject.
# There is a subject, and specific task for the dataset.
subject_id = '001' # zero padding to account for >100 subjects in this dataset
task = 'testresteyes'
# get MNE-Python directory w/ example data
mne_data_dir = mne.get_config('MNE_DATASETS_MISC_PATH')
# There is the root directory for where we will write our data.
bids_root = op.join(mne_data_dir, 'ieegmmidb_bids')
To ensure the output path doesn’t contain any leftover files from previous tests and example runs, we simply delete it.
Warning
Do not delete directories that may contain important data!
Now we just need to specify a few iEEG details to make things work:
We need the basename of the dataset. In addition, write_raw_bids()
requires the .filenames attribute of the Raw object to be non-empty,
so since we
initialized the dataset from an array, we need to do a hack where we
temporarily save the data to disc before reading it back in.
# Now convert our data to be in a new BIDS dataset.
bids_path = BIDSPath(subject=subject_id,
task=task, acquisition="ecog", root=bids_root)
# write `raw` to BIDS and anonymize it into BrainVision format
write_raw_bids(raw, bids_path, anonymize=dict(daysback=30000),
overwrite=True)
Out:
Extracting EDF parameters from /Users/hoechenberger/mne_data/MNE-misc-data/ecog/sample_ecog.edf...
EDF file detected
Setting channel info structure...
Creating raw.info structure...
Writing '/Users/hoechenberger/mne_data/ieegmmidb_bids/README'...
References
----------
Appelhoff, S., Sanderson, M., Brooks, T., Vliet, M., Quentin, R., Holdgraf, C., Chaumon, M., Mikulan, E., Tavabi, K., Höchenberger, R., Welke, D., Brunner, C., Rockhill, A., Larson, E., Gramfort, A. and Jas, M. (2019). MNE-BIDS: Organizing electrophysiological data into the BIDS format and facilitating their analysis. Journal of Open Source Software 4: (1896). https://doi.org/10.21105/joss.01896
Holdgraf, C., Appelhoff, S., Bickel, S., Bouchard, K., D'Ambrosio, S., David, O., … Hermes, D. (2019). iEEG-BIDS, extending the Brain Imaging Data Structure specification to human intracranial electrophysiology. Scientific Data, 6, 102. https://doi.org/10.1038/s41597-019-0105-7
Writing '/Users/hoechenberger/mne_data/ieegmmidb_bids/participants.tsv'...
participant_id age sex hand
sub-001 n/a n/a n/a
Writing '/Users/hoechenberger/mne_data/ieegmmidb_bids/participants.json'...
{
"participant_id": {
"Description": "Unique participant identifier"
},
"age": {
"Description": "Age of the participant at time of testing",
"Units": "years"
},
"sex": {
"Description": "Biological sex of the participant",
"Levels": {
"F": "female",
"M": "male"
}
},
"hand": {
"Description": "Handedness of the participant",
"Levels": {
"R": "right",
"L": "left",
"A": "ambidextrous"
}
}
}
Writing electrodes file to... /Users/hoechenberger/mne_data/ieegmmidb_bids/sub-001/ieeg/sub-001_acq-ecog_electrodes.tsv
Writing coordsytem file to... /Users/hoechenberger/mne_data/ieegmmidb_bids/sub-001/ieeg/sub-001_acq-ecog_coordsystem.json
Writing '/Users/hoechenberger/mne_data/ieegmmidb_bids/sub-001/ieeg/sub-001_acq-ecog_space-fsaverage_electrodes.tsv'...
name x y z size
C1 -0.0132 -0.0654 0.0702 n/a
C2 -0.0145 -0.0594 0.0781 n/a
C3 -0.0156 -0.0518 0.0838 n/a
C4 -0.0154 -0.0453 0.0895 n/a
C5 -0.0149 -0.0385 0.094 n/a
Writing '/Users/hoechenberger/mne_data/ieegmmidb_bids/sub-001/ieeg/sub-001_acq-ecog_space-fsaverage_coordsystem.json'...
{
"iEEGCoordinateSystem": "fsaverage",
"iEEGCoordinateSystemDescription": "Defined by FreeSurfer, the MRI (surface RAS) origin is at the center of a 256\u00d7256\u00d7256 mm^3 anisotropic volume (may not be in the center of the head).",
"iEEGCoordinateUnits": "m"
}
Writing '/Users/hoechenberger/mne_data/ieegmmidb_bids/sub-001/ieeg/sub-001_task-testresteyes_acq-ecog_events.tsv'...
onset duration trial_type value sample
10.229013529411764 0.0 eeg sz start 1 10223
Writing '/Users/hoechenberger/mne_data/ieegmmidb_bids/dataset_description.json'...
{
"Name": " ",
"BIDSVersion": "1.4.0",
"DatasetType": "raw",
"Authors": [
"Please cite MNE-BIDS in your publication before removing this (citations in README)"
]
}
Writing '/Users/hoechenberger/mne_data/ieegmmidb_bids/sub-001/ieeg/sub-001_task-testresteyes_acq-ecog_ieeg.json'...
{
"TaskName": "testresteyes",
"Manufacturer": "n/a",
"PowerLineFrequency": 60,
"SamplingFrequency": 999.4121105232217,
"SoftwareFilters": "n/a",
"RecordingDuration": 21.669739411764706,
"RecordingType": "continuous",
"iEEGReference": "n/a",
"ECOGChannelCount": 78,
"SEEGChannelCount": 0,
"EEGChannelCount": 0,
"EOGChannelCount": 0,
"ECGChannelCount": 0,
"EMGChannelCount": 0,
"MiscChannelCount": 0,
"TriggerChannelCount": 0
}
Writing '/Users/hoechenberger/mne_data/ieegmmidb_bids/sub-001/ieeg/sub-001_task-testresteyes_acq-ecog_channels.tsv'...
name type units low_cutoff high_cutoff description sampling_frequency status status_description
C1 ECOG µV 0.0 499.70605526161086 Electrocorticography 999.4121105232217 good n/a
C2 ECOG µV 0.0 499.70605526161086 Electrocorticography 999.4121105232217 good n/a
C3 ECOG µV 0.0 499.70605526161086 Electrocorticography 999.4121105232217 good n/a
C4 ECOG µV 0.0 499.70605526161086 Electrocorticography 999.4121105232217 good n/a
C5 ECOG µV 0.0 499.70605526161086 Electrocorticography 999.4121105232217 good n/a
Copying data files to sub-001_task-testresteyes_acq-ecog_ieeg.edf
/Users/hoechenberger/Development/mne-bids/mne_bids/write.py:1355: RuntimeWarning: EDF/EDF+/BDF files contain two fields for recording dates.Due to file format limitations, one of these fields only supports 2-digit years. The date for that field will be set to 85 (i.e., 1985), the earliest possible date. The true anonymized date is stored in the scans.tsv file.
warn("EDF/EDF+/BDF files contain two fields for recording dates."
Writing '/Users/hoechenberger/mne_data/ieegmmidb_bids/sub-001/sub-001_scans.tsv'...
filename acq_time
ieeg/sub-001_task-testresteyes_acq-ecog_ieeg.edf 1917-11-23T12:01:01.000000Z
Wrote /Users/hoechenberger/mne_data/ieegmmidb_bids/sub-001/sub-001_scans.tsv entry with ieeg/sub-001_task-testresteyes_acq-ecog_ieeg.edf.
BIDSPath(
root: /Users/hoechenberger/mne_data/ieegmmidb_bids
datatype: ieeg
basename: sub-001_task-testresteyes_acq-ecog_ieeg.edf)
Step 3: Check and compare with standard¶
# Now we have written our BIDS directory.
print_dir_tree(bids_root)
Out:
|ieegmmidb_bids/
|--- README
|--- dataset_description.json
|--- participants.json
|--- participants.tsv
|--- sub-001/
|------ sub-001_scans.tsv
|------ ieeg/
|--------- sub-001_acq-ecog_space-fsaverage_coordsystem.json
|--------- sub-001_acq-ecog_space-fsaverage_electrodes.tsv
|--------- sub-001_task-testresteyes_acq-ecog_channels.tsv
|--------- sub-001_task-testresteyes_acq-ecog_events.tsv
|--------- sub-001_task-testresteyes_acq-ecog_ieeg.edf
|--------- sub-001_task-testresteyes_acq-ecog_ieeg.json
Step 4: Cite mne-bids¶
We can see that the appropriate citations are already written in the README. If you are preparing a manuscript, please make sure to also cite MNE-BIDS there.
Out:
References
----------
Appelhoff, S., Sanderson, M., Brooks, T., Vliet, M., Quentin, R., Holdgraf, C., Chaumon, M., Mikulan, E., Tavabi, K., Höchenberger, R., Welke, D., Brunner, C., Rockhill, A., Larson, E., Gramfort, A. and Jas, M. (2019). MNE-BIDS: Organizing electrophysiological data into the BIDS format and facilitating their analysis. Journal of Open Source Software 4: (1896). https://doi.org/10.21105/joss.01896
Holdgraf, C., Appelhoff, S., Bickel, S., Bouchard, K., D'Ambrosio, S., David, O., … Hermes, D. (2019). iEEG-BIDS, extending the Brain Imaging Data Structure specification to human intracranial electrophysiology. Scientific Data, 6, 102. https://doi.org/10.1038/s41597-019-0105-7
MNE-BIDS has created a suitable directory structure for us, and among other
meta data files, it started an events.tsv` and channels.tsv file,
and created an initial dataset_description.json file on top!
Now it’s time to manually check the BIDS directory and the meta files to add
all the information that MNE-BIDS could not infer. For instance, you must
describe iEEGReference and iEEGGround yourself.
It’s easy to find these by searching for "n/a" in the sidecar files.
$ grep -i 'n/a' <bids_root>`
Remember that there is a convenient JavaScript tool to validate all your BIDS directories called the “BIDS-validator”, available as a web version and a command line tool:
Web version: https://bids-standard.github.io/bids-validator/
Command line tool: https://www.npmjs.com/package/bids-validator
Step 5: Plot output channels and check that they match!¶
Now we have written our BIDS directory. We can use
read_raw_bids() to read in the data.
# read in the BIDS dataset and plot the coordinates
raw = read_raw_bids(bids_path=bids_path)
# get the first 5 channels and show their locations
# this should match what was printed earlier.
picks = mne.pick_types(raw.info, ecog=True)
dig = [raw.info['dig'][pick] for pick in picks]
chs = [raw.info['chs'][pick] for pick in picks]
pos = np.array([ch['r'] for ch in dig[:5]])
ch_names = np.array([ch['ch_name'] for ch in chs[:5]])
print("The channel montage after writing into BIDS: ")
pprint(dig[0:5])
print("The channel coordinates after writing into BIDS: ")
pprint([x for x in zip(ch_names, pos)])
# make a plot of the sensors in 2D plane
raw.plot_sensors(ch_type='ecog')
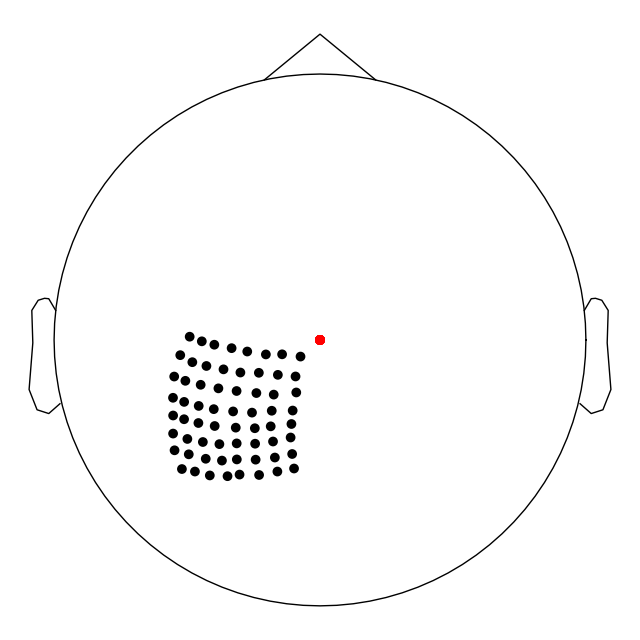
Out:
Extracting EDF parameters from /Users/hoechenberger/mne_data/ieegmmidb_bids/sub-001/ieeg/sub-001_task-testresteyes_acq-ecog_ieeg.edf...
EDF file detected
Setting channel info structure...
Creating raw.info structure...
Reading events from /Users/hoechenberger/mne_data/ieegmmidb_bids/sub-001/ieeg/sub-001_task-testresteyes_acq-ecog_events.tsv.
Reading channel info from /Users/hoechenberger/mne_data/ieegmmidb_bids/sub-001/ieeg/sub-001_task-testresteyes_acq-ecog_channels.tsv.
Reading in coordinate system frame fsaverage: None.
Reading electrode coords from /Users/hoechenberger/mne_data/ieegmmidb_bids/sub-001/ieeg/sub-001_acq-ecog_space-fsaverage_electrodes.tsv.
The read in electrodes file is:
[('name', ['C1', 'C2', 'C3', 'C4', 'C5', 'C6', 'C7', 'C8', 'C9', 'C10', 'C11', 'C12', 'C13', 'C14', 'C15', 'C16', 'C17', 'C18', 'C19', 'C20', 'C21', 'C22', 'C23', 'C24', 'C25', 'C26', 'C27', 'C28', 'C29', 'C30', 'C31', 'C32', 'C33', 'C34', 'C35', 'C36', 'C37', 'C38', 'C39', 'C40', 'C41', 'C42', 'C43', 'C44', 'C45', 'C46', 'C47', 'C48', 'C49', 'C50', 'C51', 'C52', 'C53', 'C54', 'C55', 'C56', 'C57', 'C58', 'C59', 'C60', 'C61', 'C62', 'C63', 'C64', 'BTM1', 'BTM2', 'BTM3', 'BTM4', 'BTM5', 'BTM6', 'BTP1', 'BTP2', 'BTP3', 'BTP4', 'BTP5', 'BTP6', 'EKGL', 'EKGR']), ('x', ['-0.0132', '-0.0145', '-0.0156', '-0.0154', '-0.0149', '-0.0131', '-0.0136', '-0.0109', '-0.0215', '-0.0233', '-0.0247', '-0.0263', '-0.0261', '-0.0253', '-0.0233', '-0.0211', '-0.0304', '-0.0328', '-0.0337', '-0.0344', '-0.0363', '-0.0345', '-0.0335', '-0.0299', '-0.0395', '-0.0417', '-0.0426', '-0.0438', '-0.0457', '-0.0446', '-0.043', '-0.0396', '-0.0446', '-0.0482', '-0.0505', '-0.0536', '-0.0548', '-0.0533', '-0.0512', '-0.0473', '-0.0518', '-0.055', '-0.0575', '-0.0606', '-0.0614', '-0.0612', '-0.0589', '-0.0553', '-0.0575', '-0.0615', '-0.0635', '-0.066', '-0.0671', '-0.0672', '-0.0643', '-0.0602', '-0.0614', '-0.0659', '-0.0681', '-0.0694', '-0.0704', '-0.0705', '-0.068', '-0.0642', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a']), ('y', ['-0.0654', '-0.0594', '-0.0518', '-0.0453', '-0.0385', '-0.029', '-0.0203', '-0.0093', '-0.0664', '-0.0607', '-0.0534', '-0.0461', '-0.0383', '-0.0299', '-0.0193', '-0.008', '-0.0674', '-0.061', '-0.0539', '-0.0466', '-0.0388', '-0.0288', '-0.018', '-0.008', '-0.0661', '-0.0599', '-0.0529', '-0.0456', '-0.0376', '-0.0273', '-0.0176', '-0.0063', '-0.0657', '-0.0593', '-0.0523', '-0.0438', '-0.0357', '-0.0254', '-0.0156', '-0.0044', '-0.0637', '-0.0572', '-0.0501', '-0.0414', '-0.0334', '-0.023', '-0.0135', '-0.0025', '-0.0605', '-0.0536', '-0.0474', '-0.0385', '-0.0306', '-0.0204', '-0.0112', '-0.0007', '-0.0574', '-0.05', '-0.0434', '-0.0357', '-0.0277', '-0.0177', '-0.0074', '0.0016', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a']), ('z', ['0.0702', '0.0781', '0.0838', '0.0895', '0.094', '0.0985', '0.1009', '0.1035', '0.0691', '0.0769', '0.0822', '0.0871', '0.0918', '0.0949', '0.0989', '0.1026', '0.0678', '0.0738', '0.0793', '0.0842', '0.0878', '0.0926', '0.0955', '0.0977', '0.0632', '0.069', '0.0744', '0.0795', '0.083', '0.0881', '0.0901', '0.0884', '0.0581', '0.0632', '0.0694', '0.0731', '0.0769', '0.0808', '0.0823', '0.0808', '0.0513', '0.0574', '0.0625', '0.0666', '0.0709', '0.0733', '0.0743', '0.0739', '0.0455', '0.05', '0.0554', '0.0584', '0.063', '0.0644', '0.0638', '0.064', '0.0372', '0.0412', '0.0463', '0.0503', '0.053', '0.0541', '0.053', '0.0539', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a']), ('size', ['n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a', 'n/a'])]
/Users/hoechenberger/Development/mne-bids/mne_bids/dig.py:519: RuntimeWarning: Fiducial point nasion not found, assuming identity unknown to head transformation
raw.set_montage(montage, on_missing='warn', verbose=verbose)
The channel montage after writing into BIDS:
[<DigPoint | EEG #1 : (-13.2, -65.4, 70.2) mm : MNI Talairach frame>,
<DigPoint | EEG #2 : (-14.5, -59.4, 78.1) mm : MNI Talairach frame>,
<DigPoint | EEG #3 : (-15.6, -51.8, 83.8) mm : MNI Talairach frame>,
<DigPoint | EEG #4 : (-15.4, -45.3, 89.5) mm : MNI Talairach frame>,
<DigPoint | EEG #5 : (-14.9, -38.5, 94.0) mm : MNI Talairach frame>]
The channel coordinates after writing into BIDS:
[('C1', array([-0.0132, -0.0654, 0.0702])),
('C2', array([-0.0145, -0.0594, 0.0781])),
('C3', array([-0.0156, -0.0518, 0.0838])),
('C4', array([-0.0154, -0.0453, 0.0895])),
('C5', array([-0.0149, -0.0385, 0.094 ]))]
<Figure size 640x640 with 1 Axes>
Total running time of the script: ( 0 minutes 3.964 seconds)