Note
Click here to download the full example code
Working with sensor locations¶
This tutorial describes how to read and plot sensor locations, and how the physical location of sensors is handled in MNE-Python.
Page contents
As usual we’ll start by importing the modules we need and loading some example data:
import os
import numpy as np
import matplotlib.pyplot as plt
from mpl_toolkits.mplot3d import Axes3D # noqa
import mne
sample_data_folder = mne.datasets.sample.data_path()
sample_data_raw_file = os.path.join(sample_data_folder, 'MEG', 'sample',
'sample_audvis_raw.fif')
raw = mne.io.read_raw_fif(sample_data_raw_file, preload=True, verbose=False)
About montages and layouts¶
Montages contain sensor
positions in 3D (x, y, z, in meters), and can be used to set
the physical positions of sensors. By specifying the location of sensors
relative to the brain, Montages play an
important role in computing the forward solution and computing inverse
estimates.
In contrast, Layouts are idealized 2-D
representations of sensor positions, and are primarily used for arranging
individual sensor subplots in a topoplot, or for showing the approximate
relative arrangement of sensors as seen from above.
Working with built-in montages¶
The 3D coordinates of MEG sensors are included in the raw recordings from MEG
systems, and are automatically stored in the info attribute of the
Raw file upon loading. EEG electrode locations are much more
variable because of differences in head shape. Idealized montages for many
EEG systems are included during MNE-Python installation; these files are
stored in your mne-python directory, in the
mne/channels/data/montages folder:
montage_dir = os.path.join(os.path.dirname(mne.__file__),
'channels', 'data', 'montages')
print('\nBUILT-IN MONTAGE FILES')
print('======================')
print(sorted(os.listdir(montage_dir)))
Out:
BUILT-IN MONTAGE FILES
======================
['EGI_256.csd', 'GSN-HydroCel-128.sfp', 'GSN-HydroCel-129.sfp', 'GSN-HydroCel-256.sfp', 'GSN-HydroCel-257.sfp', 'GSN-HydroCel-32.sfp', 'GSN-HydroCel-64_1.0.sfp', 'GSN-HydroCel-65_1.0.sfp', 'biosemi128.txt', 'biosemi16.txt', 'biosemi160.txt', 'biosemi256.txt', 'biosemi32.txt', 'biosemi64.txt', 'easycap-M1.txt', 'easycap-M10.txt', 'mgh60.elc', 'mgh70.elc', 'standard_1005.elc', 'standard_1020.elc', 'standard_alphabetic.elc', 'standard_postfixed.elc', 'standard_prefixed.elc', 'standard_primed.elc']
These built-in EEG montages can be loaded via
mne.channels.make_standard_montage(). Note that when loading via
make_standard_montage(), provide the filename without
its file extension:
ten_twenty_montage = mne.channels.make_standard_montage('standard_1020')
print(ten_twenty_montage)
Out:
<DigMontage | 0 extras (headshape), 0 HPIs, 3 fiducials, 94 channels>
Once loaded, a montage can be applied to data via one of the instance methods
such as raw.set_montage. It is also possible
to skip the loading step by passing the filename string directly to the
set_montage() method. This won’t work with our sample
data, because it’s channel names don’t match the channel names in the
standard 10-20 montage, so these commands are not run here:
# these will be equivalent:
# raw_1020 = raw.copy().set_montage(ten_twenty_montage)
# raw_1020 = raw.copy().set_montage('standard_1020')
Montage objects have a
plot() method for visualization of the sensor
locations in 3D; 2D projections are also possible by passing
kind='topomap':
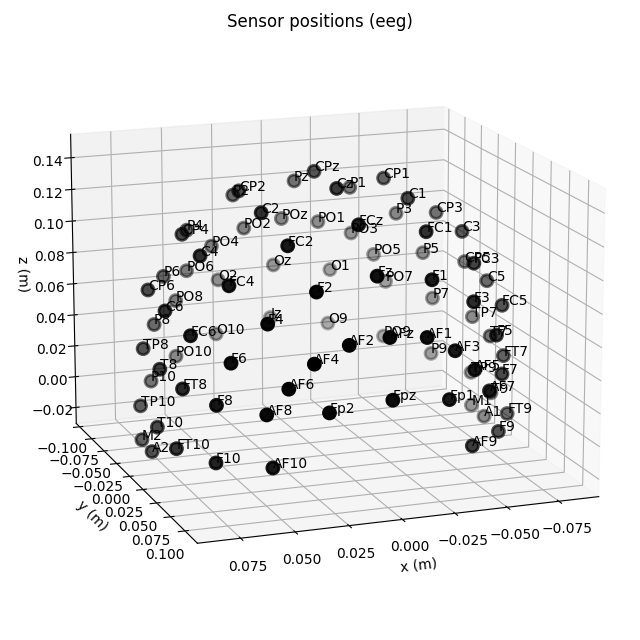
fig = ten_twenty_montage.plot(kind='3d')
fig.gca().view_init(azim=70, elev=15)
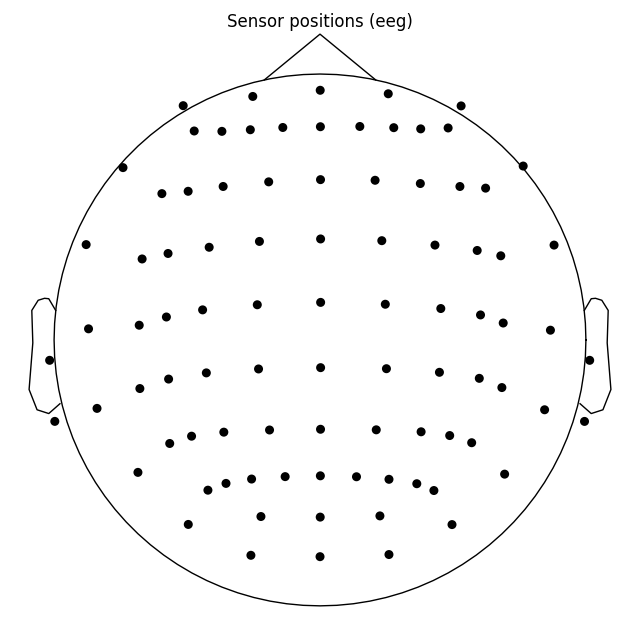
ten_twenty_montage.plot(kind='topomap', show_names=False)
Out:
4 duplicate electrode labels found:
T7/T3, T8/T4, P7/T5, P8/T6
Plotting 90 unique labels.
Creating RawArray with float64 data, n_channels=90, n_times=1
Range : 0 ... 0 = 0.000 ... 0.000 secs
Ready.
4 duplicate electrode labels found:
T7/T3, T8/T4, P7/T5, P8/T6
Plotting 90 unique labels.
Creating RawArray with float64 data, n_channels=90, n_times=1
Range : 0 ... 0 = 0.000 ... 0.000 secs
Ready.
Controlling channel projection (MNE vs EEGLAB)¶
Channel positions in 2d space are obtained by projecting their actual 3d
positions using a sphere as a reference. Because 'standard_1020' montage
contains realistic, not spherical, channel positions, we will use a different
montage to demonstrate controlling how channels are projected to 2d space.
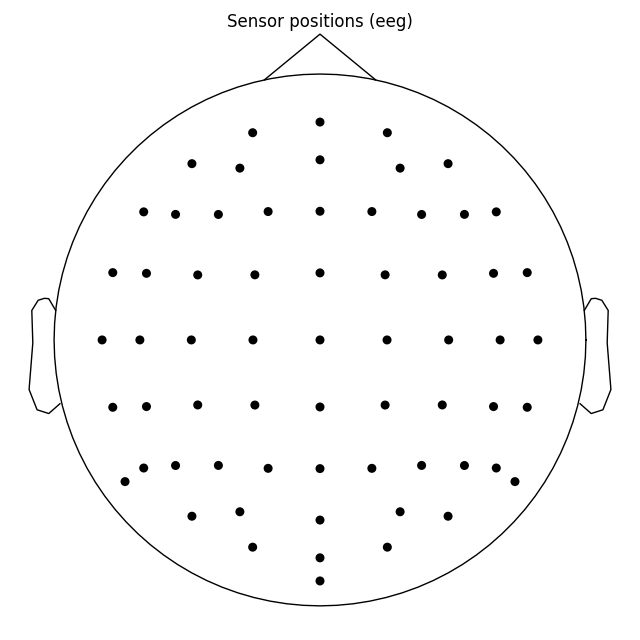
biosemi_montage = mne.channels.make_standard_montage('biosemi64')
biosemi_montage.plot(show_names=False)
Out:
Creating RawArray with float64 data, n_channels=64, n_times=1
Range : 0 ... 0 = 0.000 ... 0.000 secs
Ready.
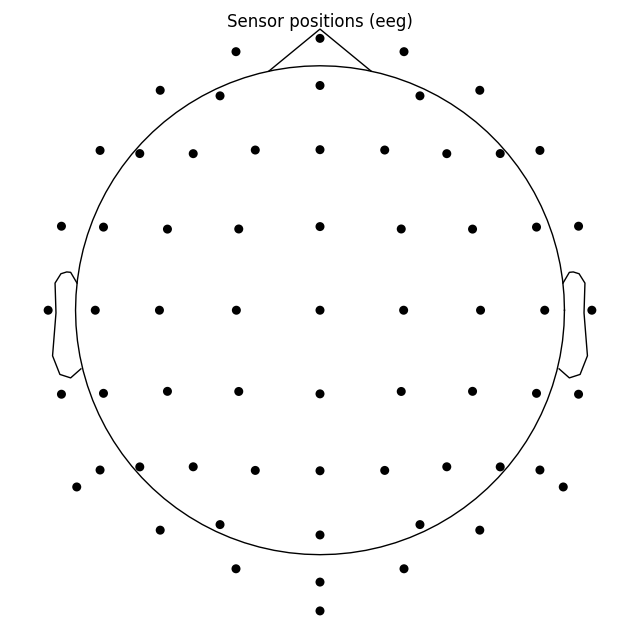
By default a sphere with an origin in (0, 0, 0) x, y, z coordinates and
radius of 0.095 meters (9.5 cm) is used. You can use a different sphere
radius by passing a single value to sphere argument in any function that
plots channels in 2d (like plot() that we use
here, but also for example mne.viz.plot_topomap()):
biosemi_montage.plot(show_names=False, sphere=0.07)
Out:
Creating RawArray with float64 data, n_channels=64, n_times=1
Range : 0 ... 0 = 0.000 ... 0.000 secs
Ready.
To control not only radius, but also the sphere origin, pass a
(x, y, z, radius) tuple to sphere argument:
biosemi_montage.plot(show_names=False, sphere=(0.03, 0.02, 0.01, 0.075))
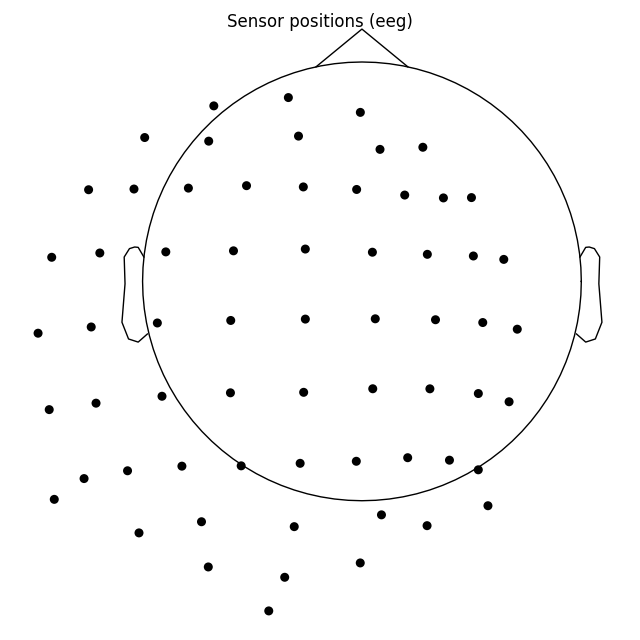
Out:
Creating RawArray with float64 data, n_channels=64, n_times=1
Range : 0 ... 0 = 0.000 ... 0.000 secs
Ready.
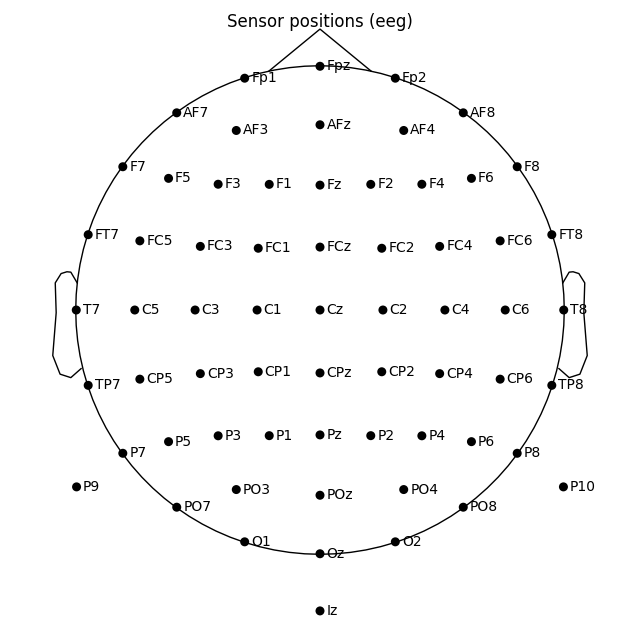
In mne-python the head center and therefore the sphere center are calculated using fiducial points. Because of this the head circle represents head circumference at the nasion and ear level, and not where it is commonly measured in 10-20 EEG system: above nasion at T4/T8, T3/T7, Oz, Fz level. Notice below that by default T7 and Oz channels are placed within the head circle, not on the head outline:
Out:
Creating RawArray with float64 data, n_channels=64, n_times=1
Range : 0 ... 0 = 0.000 ... 0.000 secs
Ready.
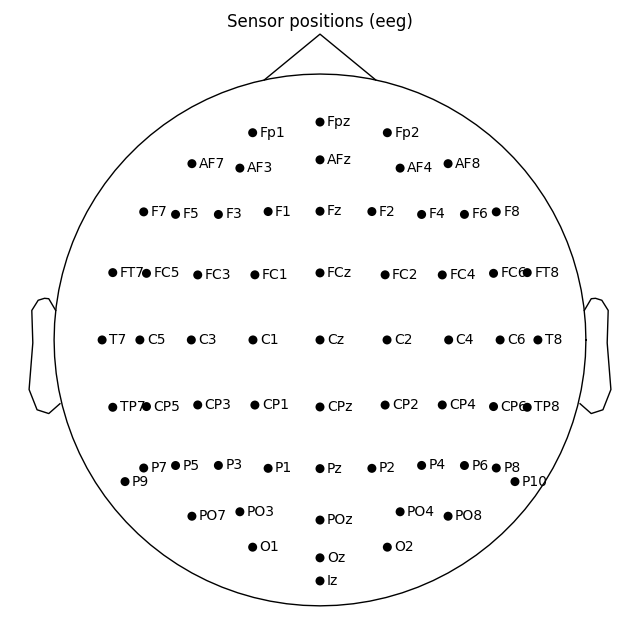
If you have previous EEGLAB experience you may prefer its convention to represent 10-20 head circumference with the head circle. To get EEGLAB-like channel layout you would have to move the sphere origin a few centimeters up on the z dimension:
biosemi_montage.plot(sphere=(0, 0, 0.035, 0.094))
Out:
Creating RawArray with float64 data, n_channels=64, n_times=1
Range : 0 ... 0 = 0.000 ... 0.000 secs
Ready.
Instead of approximating the EEGLAB-esque sphere location as above, you can calculate the sphere origin from position of Oz, Fpz, T3/T7 or T4/T8 channels. This is easier once the montage has been applied to the data and channel positions are in the head space - see this example.
Reading sensor digitization files¶
In the sample data, setting the digitized EEG montage was done prior to
saving the Raw object to disk, so the sensor positions are
already incorporated into the info attribute of the Raw
object (see the documentation of the reading functions and
set_montage() for details on how that works). Because of
that, we can plot sensor locations directly from the Raw
object using the plot_sensors() method, which provides
similar functionality to
montage.plot().
plot_sensors() also allows channel selection by type, can
color-code channels in various ways (by default, channels listed in
raw.info['bads'] will be plotted in red), and allows drawing into an
existing matplotlib axes object (so the channel positions can easily be
made as a subplot in a multi-panel figure):
fig = plt.figure()
ax2d = fig.add_subplot(121)
ax3d = fig.add_subplot(122, projection='3d')
raw.plot_sensors(ch_type='eeg', axes=ax2d)
raw.plot_sensors(ch_type='eeg', axes=ax3d, kind='3d')
ax3d.view_init(azim=70, elev=15)
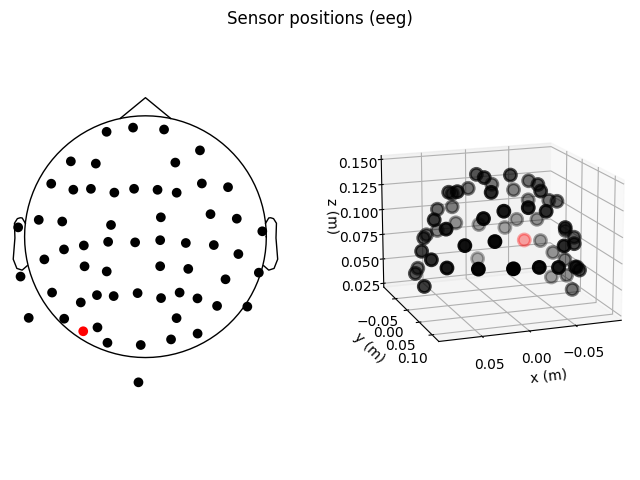
It’s probably evident from the 2D topomap above that there is some
irregularity in the EEG sensor positions in the sample dataset — this is because the sensor positions in that dataset are
digitizations of the sensor positions on an actual subject’s head, rather
than idealized sensor positions based on a spherical head model. Depending on
what system was used to digitize the electrode positions (e.g., a Polhemus
Fastrak digitizer), you must use different montage reading functions (see
Supported formats for digitized 3D locations). The resulting montage
can then be added to Raw objects by passing it to the
set_montage() method (just as we did above with the name of
the idealized montage 'standard_1020'). Once loaded, locations can be
plotted with plot() and saved with
save(), like when working with a standard
montage.
Note
When setting a montage with set_montage()
the measurement info is updated in two places (the chs
and dig entries are updated). See The Info data structure.
dig may contain HPI, fiducial, or head shape points in
addition to electrode locations.
Rendering sensor position with mayavi¶
It is also possible to render an image of a MEG sensor helmet in 3D, using
mayavi instead of matplotlib, by calling the mne.viz.plot_alignment()
function:
fig = mne.viz.plot_alignment(raw.info, trans=None, dig=False, eeg=False,
surfaces=[], meg=['helmet', 'sensors'],
coord_frame='meg')
mne.viz.set_3d_view(fig, azimuth=50, elevation=90, distance=0.5)

Out:
Getting helmet for system 306m
plot_alignment() requires an Info object, and
can also render MRI surfaces of the scalp, skull, and brain (by passing
keywords like 'head', 'outer_skull', or 'brain' to the
surfaces parameter) making it useful for assessing coordinate frame
transformations. For examples of various uses of
plot_alignment(), see Plotting sensor layouts of EEG systems,
Plotting EEG sensors on the scalp, and
Plotting sensor layouts of MEG systems.
Working with layout files¶
As with montages, many layout files are included during MNE-Python
installation, and are stored in the mne/channels/data/layouts folder:
layout_dir = os.path.join(os.path.dirname(mne.__file__),
'channels', 'data', 'layouts')
print('\nBUILT-IN LAYOUT FILES')
print('=====================')
print(sorted(os.listdir(layout_dir)))
Out:
BUILT-IN LAYOUT FILES
=====================
['CTF-275.lout', 'CTF151.lay', 'CTF275.lay', 'EEG1005.lay', 'EGI256.lout', 'KIT-125.lout', 'KIT-157.lout', 'KIT-160.lay', 'KIT-AD.lout', 'KIT-AS-2008.lout', 'KIT-UMD-3.lout', 'Neuromag_122.lout', 'Vectorview-all.lout', 'Vectorview-grad.lout', 'Vectorview-grad_norm.lout', 'Vectorview-mag.lout', 'biosemi.lay', 'magnesWH3600.lout']
You may have noticed that the file formats and filename extensions of the built-in layout and montage files vary considerably. This reflects different manufacturers’ conventions; to make loading easier the montage and layout loading functions in MNE-Python take the filename without its extension so you don’t have to keep track of which file format is used by which manufacturer.
To load a layout file, use the mne.channels.read_layout() function, and
provide the filename without its file extension. You can then visualize the
layout using its plot() method, or (equivalently)
by passing it to mne.viz.plot_layout():
biosemi_layout = mne.channels.read_layout('biosemi')
biosemi_layout.plot() # same result as: mne.viz.plot_layout(biosemi_layout)
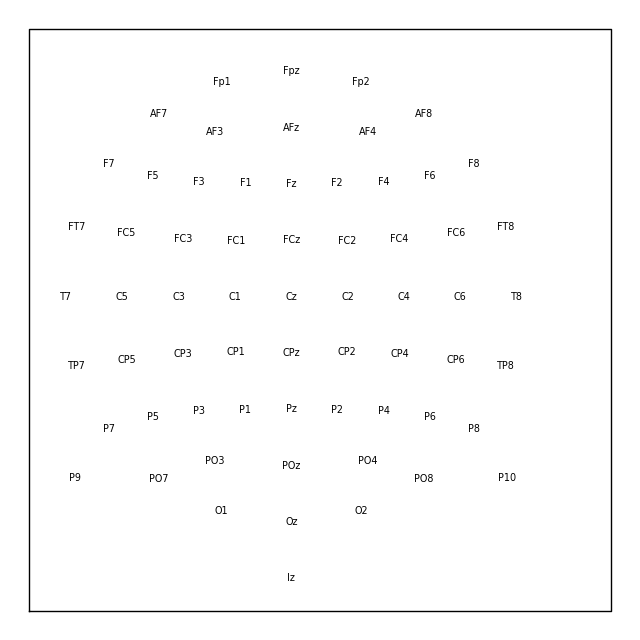
Similar to the picks argument for selecting channels from
Raw objects, the plot() method of
Layout objects also has a picks argument. However,
because layouts only contain information about sensor name and location (not
sensor type), the plot() method only allows
picking channels by index (not by name or by type). Here we find the indices
we want using numpy.where(); selection by name or type is possible via
mne.pick_channels() or mne.pick_types().
midline = np.where([name.endswith('z') for name in biosemi_layout.names])[0]
biosemi_layout.plot(picks=midline)
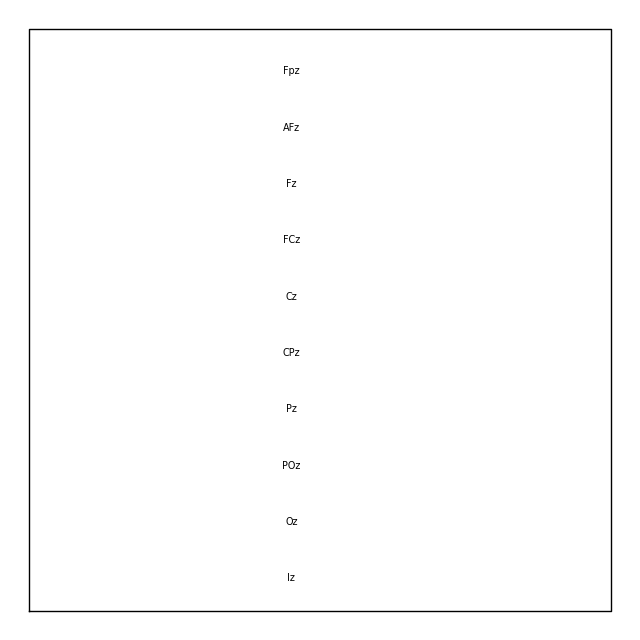
If you’re working with a Raw object that already has sensor
positions incorporated, you can create a Layout object
with either the mne.channels.make_eeg_layout() function or
(equivalently) the mne.channels.find_layout() function.
layout_from_raw = mne.channels.make_eeg_layout(raw.info)
# same result as: mne.channels.find_layout(raw.info, ch_type='eeg')
layout_from_raw.plot()

Note
There is no corresponding make_meg_layout function because sensor
locations are fixed in a MEG system (unlike in EEG, where the sensor caps
deform to fit each subject’s head). Thus MEG layouts are consistent for a
given system and you can simply load them with
mne.channels.read_layout(), or use mne.channels.find_layout()
with the ch_type parameter, as shown above for EEG.
All Layout objects have a
save() method that allows writing layouts to disk,
in either .lout or .lay format (which format gets written is
inferred from the file extension you pass to the method’s fname
parameter). The choice between .lout and .lay format only
matters if you need to load the layout file in some other software
(MNE-Python can read either format equally well).
Total running time of the script: ( 0 minutes 24.444 seconds)
Estimated memory usage: 492 MB