mne.viz.plot_alignment¶
-
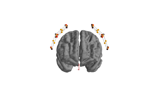
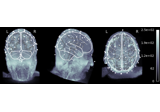
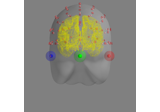
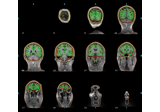
mne.viz.plot_alignment(info=None, trans=None, subject=None, subjects_dir=None, surfaces='auto', coord_frame='head', meg=None, eeg='original', fwd=None, dig=False, ecog=True, src=None, mri_fiducials=False, bem=None, seeg=True, fnirs=True, show_axes=False, fig=None, interaction='trackball', verbose=None)[source]¶ Plot head, sensor, and source space alignment in 3D.
- Parameters
- info
dict|None The measurement info. If None (default), no sensor information will be shown.
- trans
str|dict| instance ofTransform|None If str, the path to the head<->MRI transform
*-trans.fiffile produced during coregistration. Can also be'fsaverage'to use the built-in fsaverage transformation. If trans is None, an identity matrix is assumed.Changed in version 0.19: Support for ‘fsaverage’ argument.
- subject
str|None The subject name corresponding to FreeSurfer environment variable SUBJECT. Can be omitted if
srcis provided.- subjects_dir
str|None The path to the freesurfer subjects reconstructions. It corresponds to Freesurfer environment variable SUBJECTS_DIR.
- surfaces
str|list Surfaces to plot. Supported values:
scalp: one of ‘head’, ‘outer_skin’ (alias for ‘head’), ‘head-dense’, or ‘seghead’ (alias for ‘head-dense’)
skull: ‘outer_skull’, ‘inner_skull’, ‘brain’ (alias for ‘inner_skull’)
brain: one of ‘pial’, ‘white’, ‘inflated’, or ‘brain’ (alias for ‘pial’).
Defaults to ‘auto’, which will look for a head surface and plot it if found.
Note
For single layer BEMs it is recommended to use ‘brain’.
- coord_frame
str Coordinate frame to use, ‘head’, ‘meg’, or ‘mri’.
- meg
str|list| bool |None Can be “helmet”, “sensors” or “ref” to show the MEG helmet, sensors or reference sensors respectively, or a combination like
('helmet', 'sensors')(same as None, default). True translates to('helmet', 'sensors', 'ref').- eegbool |
str|list String options are:
- “original” (default; equivalent to
True) Shows EEG sensors using their digitized locations (after transformation to the chosen
coord_frame)
- “original” (default; equivalent to
- “projected”
The EEG locations projected onto the scalp, as is done in forward modeling
Can also be a list of these options, or an empty list (
[], equivalent ofFalse).- fwdinstance of
Forward The forward solution. If present, the orientations of the dipoles present in the forward solution are displayed.
- digbool | ‘fiducials’
If True, plot the digitization points; ‘fiducials’ to plot fiducial points only.
- ecogbool
If True (default), show ECoG sensors.
- srcinstance of
SourceSpaces|None If not None, also plot the source space points.
- mri_fiducialsbool |
str Plot MRI fiducials (default False). If
True, look for a file with the canonical name (bem/{subject}-fiducials.fif). Ifstrit should provide the full path to the fiducials file.- bem
listofdict| instance ofConductorModel|None Can be either the BEM surfaces (list of dict), a BEM solution or a sphere model. If None, we first try loading
'$SUBJECTS_DIR/$SUBJECT/bem/$SUBJECT-$SOURCE.fif', and then look for'$SUBJECT*$SOURCE.fif'in the same directory. For'outer_skin', the subjects bem and bem/flash folders are searched. Defaults to None.- seegbool
If True (default), show sEEG electrodes.
- fnirs
str|list| bool |None Can be “channels”, “pairs”, “detectors”, and/or “sources” to show the fNIRS channel locations, optode locations, or line between source-detector pairs, or a combination like
('pairs', 'channels'). True translates to('pairs',).New in version 0.20.
- show_axesbool
If True (default False), coordinate frame axis indicators will be shown:
head in pink.
MRI in gray (if
trans is not None).MEG in blue (if MEG sensors are present).
New in version 0.16.
- fig
mayavi.mlab.Figure|None Mayavi Scene in which to plot the alignment. If
None, creates a new 600x600 pixel figure with black background.New in version 0.16.
- interaction
str Can be “trackball” (default) or “terrain”, i.e. a turntable-style camera.
New in version 0.16.
- verbosebool,
str,int, orNone If not None, override default verbose level (see
mne.verbose()and Logging documentation for more). If used, it should be passed as a keyword-argument only.
- info
- Returns
- figinstance of
mayavi.mlab.Figure The mayavi figure.
- figinstance of
See also
Notes
This function serves the purpose of checking the validity of the many different steps of source reconstruction:
Transform matrix (keywords
trans,megandmri_fiducials),BEM surfaces (keywords
bemandsurfaces),sphere conductor model (keywords
bemandsurfaces) andsource space (keywords
surfacesandsrc).
New in version 0.15.