Note
Click here to download the full example code
EEG processing and Event Related Potentials (ERPs)¶
Here we cover the specifics of EEG, namely:
import matplotlib.pyplot as plt
import mne
from mne.datasets import sample
from mne.channels import combine_channels
Setup for reading the raw data
data_path = sample.data_path()
raw_fname = data_path + '/MEG/sample/sample_audvis_filt-0-40_raw.fif'
event_fname = data_path + '/MEG/sample/sample_audvis_filt-0-40_raw-eve.fif'
raw = mne.io.read_raw_fif(raw_fname)
Out:
Opening raw data file /home/circleci/mne_data/MNE-sample-data/MEG/sample/sample_audvis_filt-0-40_raw.fif...
Read a total of 4 projection items:
PCA-v1 (1 x 102) idle
PCA-v2 (1 x 102) idle
PCA-v3 (1 x 102) idle
Average EEG reference (1 x 60) idle
Range : 6450 ... 48149 = 42.956 ... 320.665 secs
Ready.
Let’s restrict the data to the EEG channels
raw.pick_types(meg=False, eeg=True, eog=True).load_data()
# This particular dataset already has an average reference projection added
# that we now want to remove it for the sake of this example.
raw.set_eeg_reference([])
Out:
Removing projector <Projection | PCA-v1, active : False, n_channels : 102>
Removing projector <Projection | PCA-v2, active : False, n_channels : 102>
Removing projector <Projection | PCA-v3, active : False, n_channels : 102>
Reading 0 ... 41699 = 0.000 ... 277.709 secs...
EEG channel type selected for re-referencing
EEG data marked as already having the desired reference.
Removing existing average EEG reference projection.
By looking at the measurement info you will see that we have now 59 EEG channels and 1 EOG channel
print(raw.info)
Out:
<Info | 13 non-empty values
bads: []
ch_names: EEG 001, EEG 002, EEG 003, EEG 004, EEG 005, EEG 006, EEG 007, ...
chs: 59 EEG, 1 EOG
custom_ref_applied: False
dev_head_t: MEG device -> head transform
dig: 146 items (3 Cardinal, 4 HPI, 61 EEG, 78 Extra)
file_id: 4 items (dict)
highpass: 0.1 Hz
hpi_meas: 1 item (list)
hpi_results: 1 item (list)
lowpass: 40.0 Hz
meas_date: 2002-12-03 19:01:10 UTC
meas_id: 4 items (dict)
nchan: 60
projs: []
sfreq: 150.2 Hz
>
In practice it’s quite common to have some EEG channels that are actually
EOG channels. To change a channel type you can use the
mne.io.Raw.set_channel_types() method. For example
to treat an EOG channel as EEG you can change its type using
raw.set_channel_types(mapping={'EOG 061': 'eeg'})
print(raw.info)
Out:
<Info | 13 non-empty values
bads: []
ch_names: EEG 001, EEG 002, EEG 003, EEG 004, EEG 005, EEG 006, EEG 007, ...
chs: 60 EEG
custom_ref_applied: False
dev_head_t: MEG device -> head transform
dig: 146 items (3 Cardinal, 4 HPI, 61 EEG, 78 Extra)
file_id: 4 items (dict)
highpass: 0.1 Hz
hpi_meas: 1 item (list)
hpi_results: 1 item (list)
lowpass: 40.0 Hz
meas_date: 2002-12-03 19:01:10 UTC
meas_id: 4 items (dict)
nchan: 60
projs: []
sfreq: 150.2 Hz
>
And to change the name of the EOG channel
raw.rename_channels(mapping={'EOG 061': 'EOG'})
Let’s reset the EOG channel back to EOG type.
raw.set_channel_types(mapping={'EOG': 'eog'})
The EEG channels in the sample dataset already have locations. These locations are available in the ‘loc’ of each channel description. For the first channel we get
print(raw.info['chs'][0]['loc'])
Out:
[-0.03737009 0.10568011 0.07333875 0.00235201 0.11096951 -0.03500458
0. 1. 0. 0. 0. 1. ]
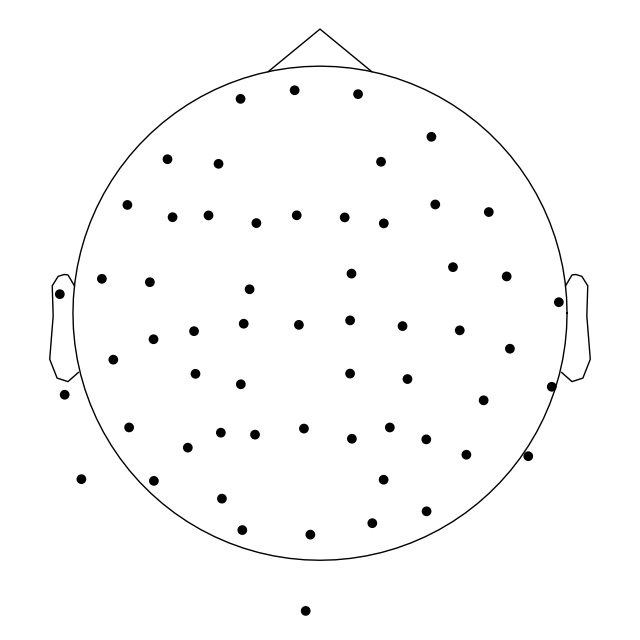
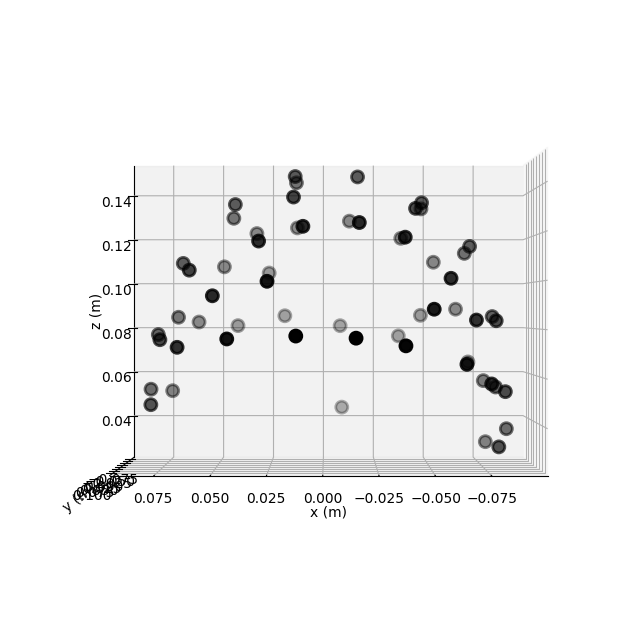
And it’s actually possible to plot the channel locations using
mne.io.Raw.plot_sensors().
In the case where your data don’t have locations you can use one of the
standard Montages shipped with MNE.
See Plotting sensor layouts of EEG systems and EEG forward operator with a template MRI.
raw.plot_sensors()
raw.plot_sensors('3d') # in 3D
Setting EEG reference¶
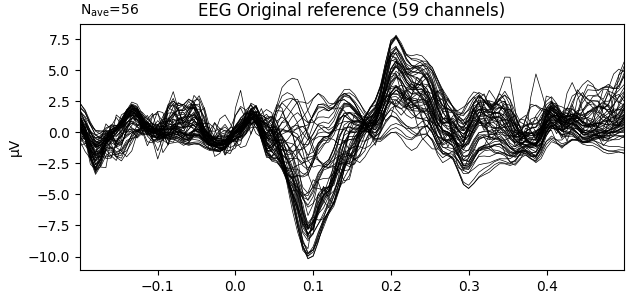
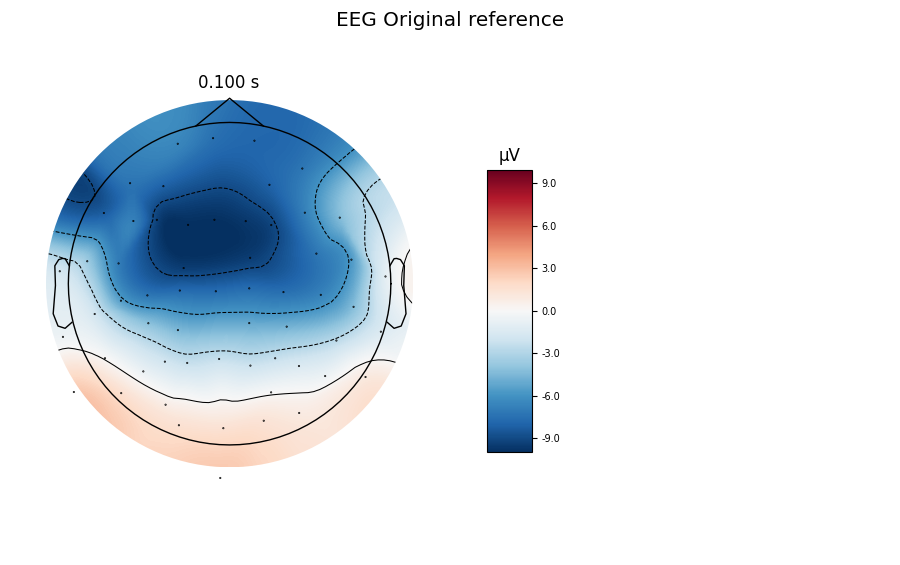
Let’s first inspect our Raw object with its original reference that was applied during the recording of the data. We define Epochs and compute an ERP for the left auditory condition.
reject = dict(eeg=180e-6, eog=150e-6)
event_id, tmin, tmax = {'left/auditory': 1}, -0.2, 0.5
events = mne.read_events(event_fname)
epochs_params = dict(events=events, event_id=event_id, tmin=tmin, tmax=tmax,
reject=reject)
evoked_no_ref = mne.Epochs(raw, **epochs_params).average()
title = 'EEG Original reference'
evoked_no_ref.plot(titles=dict(eeg=title), time_unit='s')
evoked_no_ref.plot_topomap(times=[0.1], size=3., title=title, time_unit='s')
Out:
Not setting metadata
Not setting metadata
72 matching events found
Setting baseline interval to [-0.19979521315838786, 0.0] sec
Applying baseline correction (mode: mean)
0 projection items activated
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EEG : ['EEG 007']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EEG : ['EEG 001', 'EEG 002', 'EEG 003', 'EEG 007']
Rejecting epoch based on EEG : ['EEG 001', 'EEG 002', 'EEG 003', 'EEG 007']
Common average reference (car): We add back the average reference projection that we removed at the beginning of this example (right after loading the data).
raw_car, _ = mne.set_eeg_reference(raw, 'average', projection=True)
evoked_car = mne.Epochs(raw_car, **epochs_params).average()
del raw_car # save memory
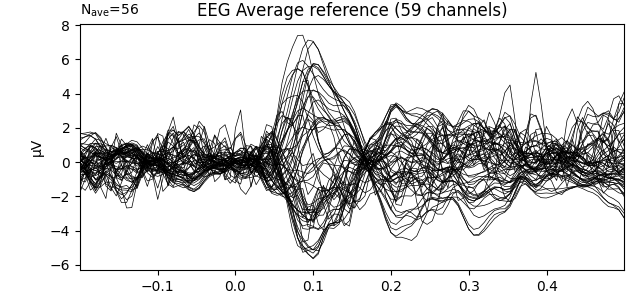
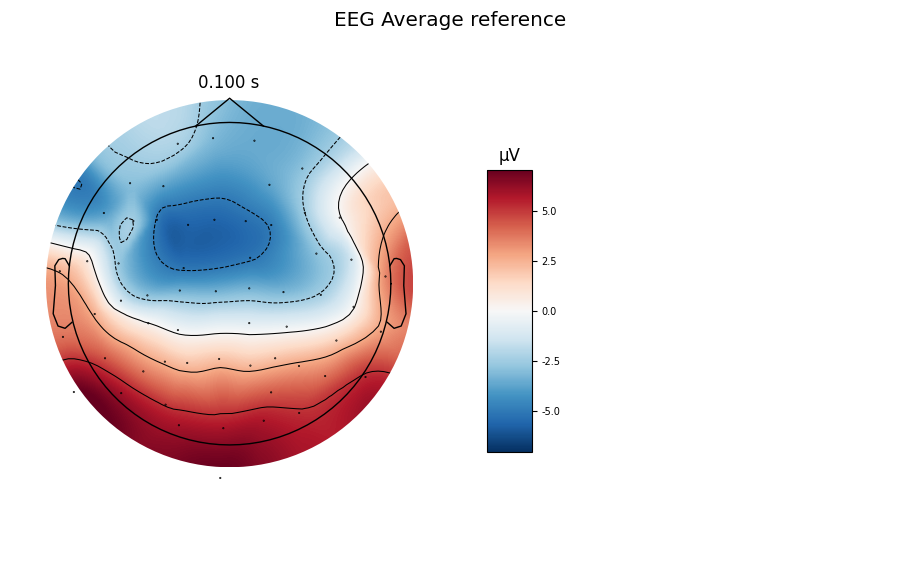
title = 'EEG Average reference'
evoked_car.plot(titles=dict(eeg=title), time_unit='s')
evoked_car.plot_topomap(times=[0.1], size=3., title=title, time_unit='s')
Out:
Adding average EEG reference projection.
1 projection items deactivated
Average reference projection was added, but has not been applied yet. Use the apply_proj method to apply it.
Not setting metadata
Not setting metadata
72 matching events found
Setting baseline interval to [-0.19979521315838786, 0.0] sec
Applying baseline correction (mode: mean)
Created an SSP operator (subspace dimension = 1)
1 projection items activated
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
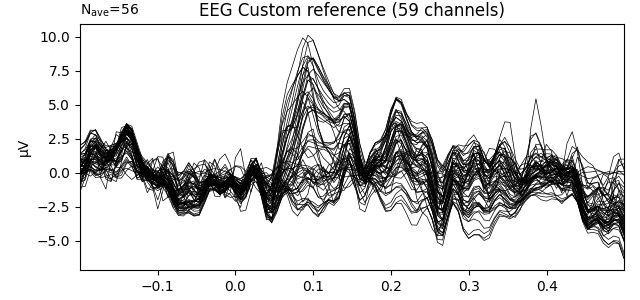
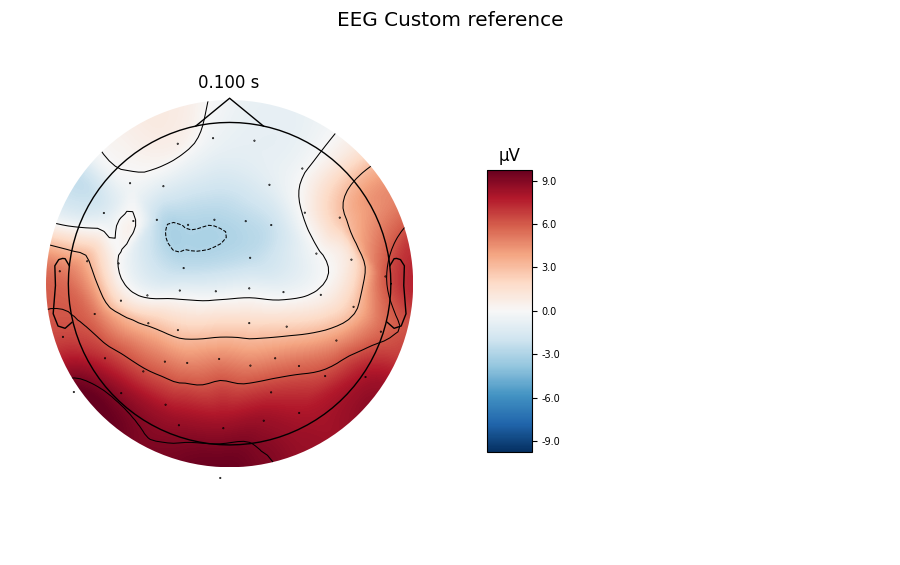
Custom reference: Use the mean of channels EEG 001 and EEG 002 as a reference
raw_custom, _ = mne.set_eeg_reference(raw, ['EEG 001', 'EEG 002'])
evoked_custom = mne.Epochs(raw_custom, **epochs_params).average()
del raw_custom # save memory
title = 'EEG Custom reference'
evoked_custom.plot(titles=dict(eeg=title), time_unit='s')
evoked_custom.plot_topomap(times=[0.1], size=3., title=title, time_unit='s')
Out:
EEG channel type selected for re-referencing
Applying a custom EEG reference.
Removing existing average EEG reference projection.
Not setting metadata
Not setting metadata
72 matching events found
Setting baseline interval to [-0.19979521315838786, 0.0] sec
Applying baseline correction (mode: mean)
0 projection items activated
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
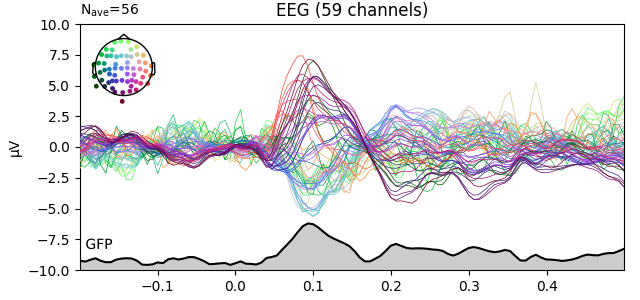
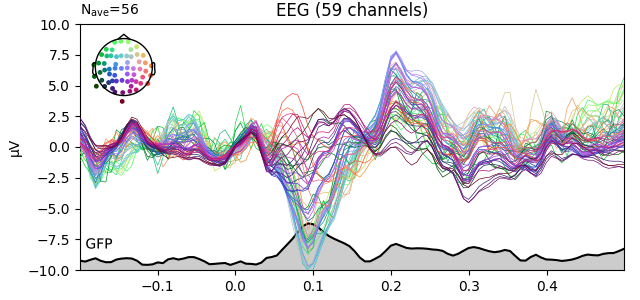
Global field power (GFP)¶
Global field power 123 is, generally speaking, a measure of agreement of the signals picked up by all sensors across the entire scalp: if all sensors have the same value at a given time point, the GFP will be zero at that time point; if the signals differ, the GFP will be non-zero at that time point. GFP peaks may reflect “interesting” brain activity, warranting further investigation. Mathematically, the GFP is the population standard deviation across all sensors, calculated separately for every time point.
You can plot the GFP using evoked.plot(gfp=True). The GFP
trace will be black if spatial_colors=True and green otherwise. The EEG
reference will not affect the GFP:
for evk in (evoked_car, evoked_no_ref):
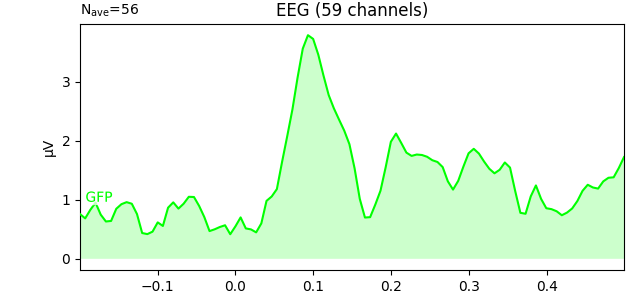
evk.plot(gfp=True, spatial_colors=True, ylim=dict(eeg=[-10, 10]))
To plot the GFP by itself you can pass gfp='only' (this makes it easier
to read off the GFP data values, because the scale is aligned):
evoked_car.plot(gfp='only')
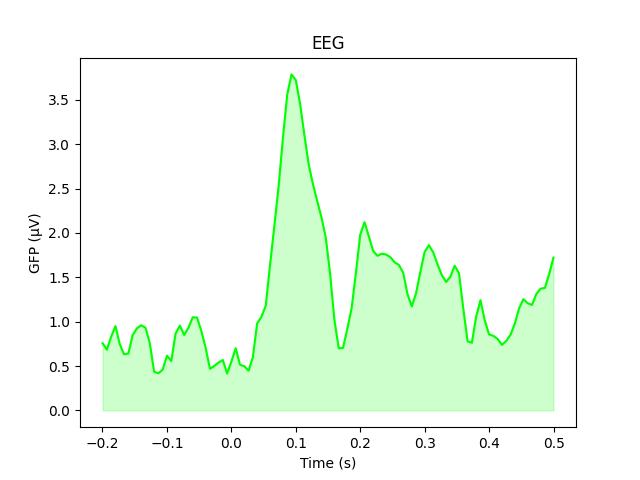
As stated above, the GFP is the population standard deviation of the signal
across channels. To compute it manually, we can leverage
the fact that evoked.data is a NumPy array:
gfp = evoked_car.data.std(axis=0, ddof=0)
# Reproducing the plot style from above:
fig, ax = plt.subplots()
ax.plot(evoked_car.times, gfp * 1e6, color='lime')
ax.fill_between(evoked_car.times, gfp * 1e6, color='lime', alpha=0.2)
ax.set(xlabel='Time (s)', ylabel='GFP (µV)', title='EEG')
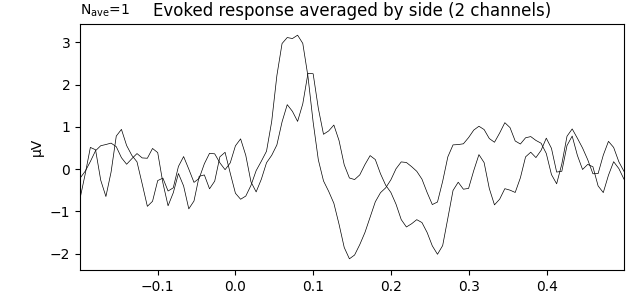
Evoked response averaged across channels by ROI¶
It is possible to average channels by region of interest (for example left and right) when studying the response to this left auditory stimulus. Here we use our Raw object on which the average reference projection has been added back.
evoked = mne.Epochs(raw, **epochs_params).average()
left_idx = mne.pick_channels(evoked.info['ch_names'],
['EEG 017', 'EEG 018', 'EEG 025', 'EEG 026'])
right_idx = mne.pick_channels(evoked.info['ch_names'],
['EEG 023', 'EEG 024', 'EEG 034', 'EEG 035'])
roi_dict = dict(Left=left_idx, Right=right_idx)
evoked_combined = combine_channels(evoked, roi_dict, method='mean')
title = 'Evoked response averaged by side'
evoked_combined.plot(titles=dict(eeg=title), time_unit='s')
Out:
Not setting metadata
Not setting metadata
72 matching events found
Setting baseline interval to [-0.19979521315838786, 0.0] sec
Applying baseline correction (mode: mean)
Created an SSP operator (subspace dimension = 1)
1 projection items activated
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Evoked arithmetic (e.g. differences)¶
Trial subsets from Epochs can be selected using ‘tags’ separated by ‘/’. Evoked objects support basic arithmetic. First, we create an Epochs object containing 4 conditions.
Out:
Not setting metadata
Not setting metadata
288 matching events found
Setting baseline interval to [-0.19979521315838786, 0.0] sec
Applying baseline correction (mode: mean)
Created an SSP operator (subspace dimension = 1)
1 projection items activated
<Epochs | 288 events (good & bad), -0.199795 - 0.499488 sec, baseline [-0.199795, 0] sec, ~2.9 MB, data not loaded,
'left/auditory': 72
'left/visual': 73
'right/auditory': 73
'right/visual': 70>
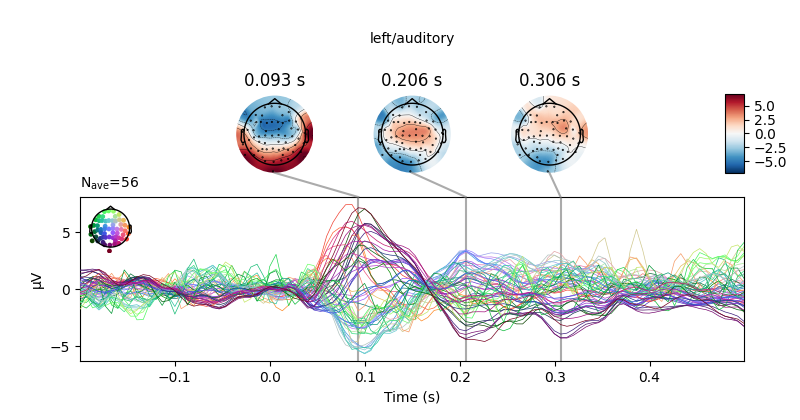
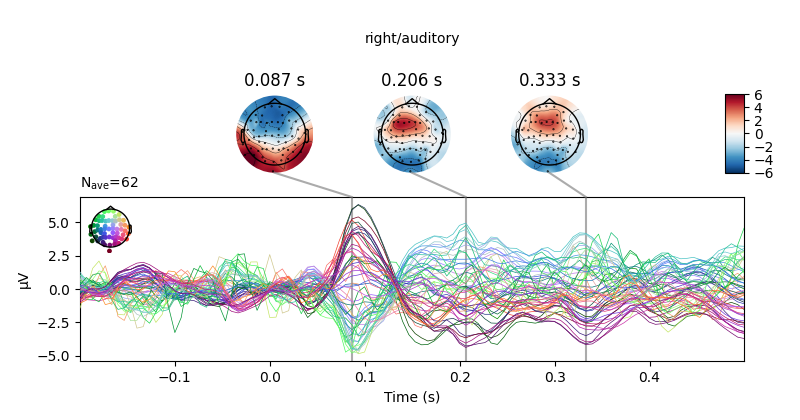
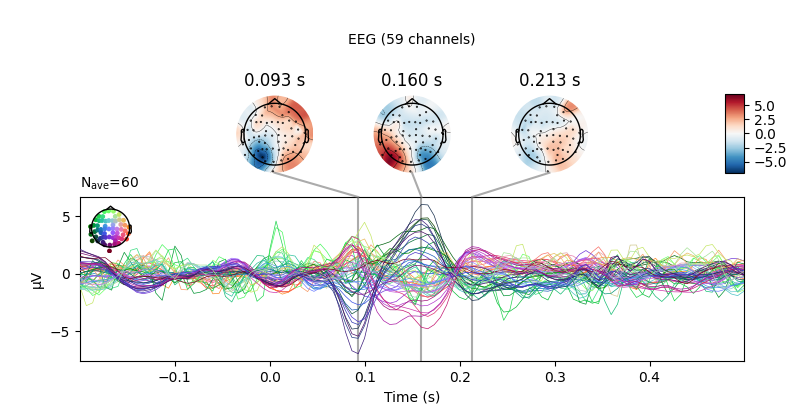
Next, we create averages of stimulation-left vs stimulation-right trials.
We can use negative weights in mne.combine_evoked to construct difference
ERPs.
left, right = epochs["left"].average(), epochs["right"].average()
# create and plot difference ERP
joint_kwargs = dict(ts_args=dict(time_unit='s'),
topomap_args=dict(time_unit='s'))
mne.combine_evoked([left, right], weights=[1, -1]).plot_joint(**joint_kwargs)
Out:
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EEG : ['EEG 001']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Projections have already been applied. Setting proj attribute to True.
This is an equal-weighting difference. If you have imbalanced trial numbers,
you could also consider either equalizing the number of events per
condition (using
epochs.equalize_event_counts) or
use weights proportional to the number of trials averaged together to create
each Evoked (by passing weights='nave' to combine_evoked).
As an example, first, we create individual ERPs for each condition.
Out:
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EEG : ['EEG 001']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
[<Evoked | 'left/auditory' (average, N=56), [-0.1998, 0.49949] sec, 59 ch, ~3.0 MB>, <Evoked | 'right/auditory' (average, N=62), [-0.1998, 0.49949] sec, 59 ch, ~3.0 MB>, <Evoked | 'left/visual' (average, N=67), [-0.1998, 0.49949] sec, 59 ch, ~3.0 MB>, <Evoked | 'right/visual' (average, N=56), [-0.1998, 0.49949] sec, 59 ch, ~3.0 MB>]
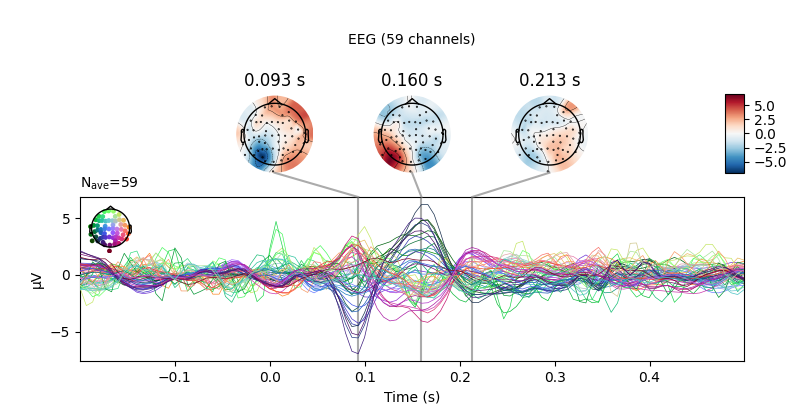
This can be simplified with a Python list comprehension:
all_evokeds = [epochs[cond].average() for cond in sorted(event_id.keys())]
print(all_evokeds)
# Then, we can construct and plot an unweighted average of left vs. right
# trials this way, too:
mne.combine_evoked(
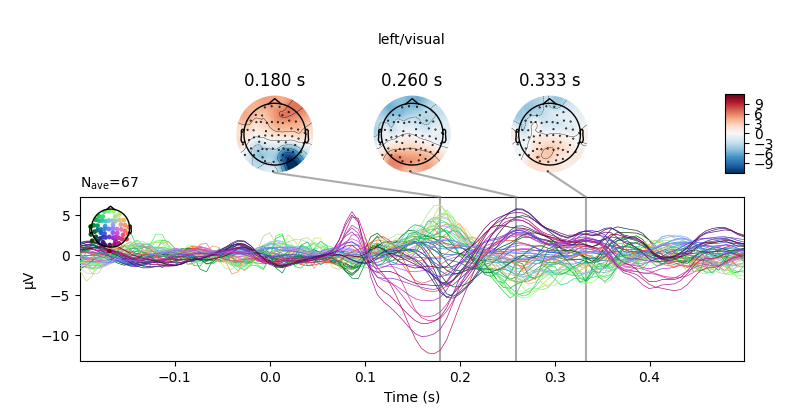
all_evokeds, weights=[0.5, 0.5, -0.5, -0.5]).plot_joint(**joint_kwargs)
Out:
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EEG : ['EEG 001']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
[<Evoked | 'left/auditory' (average, N=56), [-0.1998, 0.49949] sec, 59 ch, ~3.0 MB>, <Evoked | 'left/visual' (average, N=67), [-0.1998, 0.49949] sec, 59 ch, ~3.0 MB>, <Evoked | 'right/auditory' (average, N=62), [-0.1998, 0.49949] sec, 59 ch, ~3.0 MB>, <Evoked | 'right/visual' (average, N=56), [-0.1998, 0.49949] sec, 59 ch, ~3.0 MB>]
Projections have already been applied. Setting proj attribute to True.
Often, it makes sense to store Evoked objects in a dictionary or a list - either different conditions, or different subjects.
# If they are stored in a list, they can be easily averaged, for example,
# for a grand average across subjects (or conditions).
grand_average = mne.grand_average(all_evokeds)
# And they can be written to disk like any other evoked data, e.g.:
# mne.write_evokeds('tmp-ave.fif', all_evokeds)
# If Evokeds objects are stored in a dictionary, they can be retrieved by name.
all_evokeds = dict((cond, epochs[cond].average()) for cond in event_id)
print(all_evokeds['left/auditory'])
# Besides for explicit access, this can be used for example to set titles.
for cond in all_evokeds:
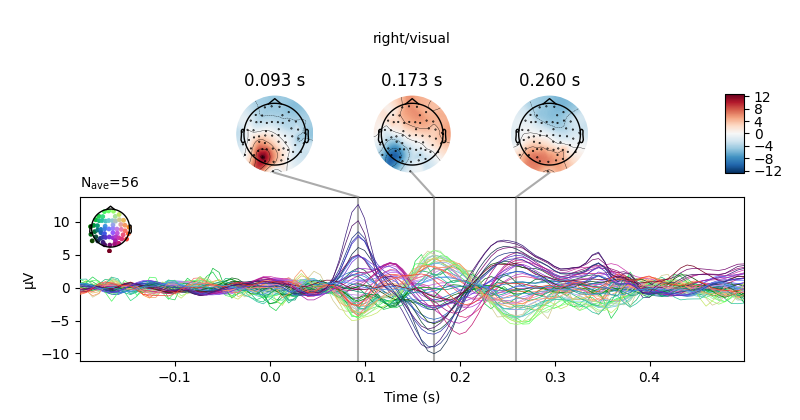
all_evokeds[cond].plot_joint(title=cond, **joint_kwargs)
Out:
Identifying common channels ...
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EEG : ['EEG 001']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
Rejecting epoch based on EOG : ['EOG']
<Evoked | 'left/auditory' (average, N=56), [-0.1998, 0.49949] sec, 59 ch, ~3.0 MB>
Projections have already been applied. Setting proj attribute to True.
Projections have already been applied. Setting proj attribute to True.
Projections have already been applied. Setting proj attribute to True.
Projections have already been applied. Setting proj attribute to True.
References¶
- 1
Dietrich Lehmann and Wolfgang Skrandies. Reference-free identification of components of checkerboard-evoked multichannel potential fields. Electroencephalography and Clinical Neurophysiology, 48(6):609–621, jun 1980. doi:10.1016/0013-4694(80)90419-8.
- 2
Dietrich Lehmann and Wolfgang Skrandies. Spatial analysis of evoked potentials in man—a review. Progress in Neurobiology, 23(3):227–250, jan 1984. doi:10.1016/0301-0082(84)90003-0.
- 3
Micah M. Murray, Denis Brunet, and Christoph M. Michel. Topographic ERP analyses: A step-by-step tutorial review. Brain Topography, 20(4):249–264, mar 2008. doi:10.1007/s10548-008-0054-5.
Total running time of the script: ( 0 minutes 26.201 seconds)
Estimated memory usage: 8 MB