Roadmap#
This page describes some of the major medium- to long-term goals for MNE-Python. These are goals that require substantial effort and/or API design considerations. Some of these may be suitable for Google Summer of Code projects, while others require more extensive work.
Open#
Type Annotations#
We would like to have type annotations for as much of our codebase as is practicable. The main motivation for this is to improve the end-user experience when writing analysis code that uses MNE-Python (i.e., code-completion suggestions, which rely on static analysis / type hints). The main discussion of how to go about this is in #12243. Some piecemeal progress has been made (e.g., #12250) but there isn’t currently anyone actively chipping away at this, hence its status as “open” rather than “in progress”.
Docstring De-duplication#
For many years, MNE-Python has used a technique borrowed from SciPy (called
doccer)
for improving the consistency of parameter names and descriptions that recur across our
API. For example, parameters for number of parallel jobs to use, for specifying random
seeds, or for controlling the appearance of a colorbar on a plot — all of these appear
in multiple functions/methods in MNE-Python. The approach works by re-defining a
function’s __doc__ attribute at import time, filling in placeholders in the
docstring’s parameter list with fully spelled-out equivalents (which are stored in a big
dictionary called the docdict). There are two major downsides:
Many docstrings can’t be read (at least not in full) while browsing the source code.
Static code analyzers don’t have access to the completed docstrings, so things like hover-tooltips in IDEs are less useful than they would be if the docstrings were complete in-place.
A possible route forward:
Convert all docstrings to be fully spelled out in the source code.
Instead of maintaining the
docdict, maintain a registry of sets of function+parameter combinations that ought to be identical.Add a test that the entries in the registry are indeed identical, so that inconsistencies cannot be introduced in existing code.
Add a test that parses docstrings in any newly added functions and looks for parameter names that maybe should be added to the registry of identical docstrings.
To allow for parameter descriptions that should be nearly identical (e.g., the same except one refers to
Rawobjects and the other refers toEpochsobjects), consider using regular expressions to check the “identity” of the parameter descriptions.
The main discussion is in #8218; a wider discussion among maintainers of other packages in the Scientific Python Ecosystem is here.
Containerization#
Users sometimes encounter difficulty getting a working MNE-Python environment on shared resources (such as compute clusters), due to various problems (old versions of package managers or graphics libraries, lack of sufficient permissions, etc). Providing a robust and up-to-date containerized distribution of MNE-Python would alleviate some of these issues. Initial efforts can be seen in the MNE-Docker repository; these efforts should be revived, brought up-to-date as necessary, and integrated into our normal release process so that the images do not become stale.
Education#
Live workshops/tutorials/trainings on MNE-Python have historically been organized ad-hoc rather than centrally. Instructors for these workshops are often approached directly by the organization or group desiring to host the training, and there is often no way for users outside that group to attend (or even learn about the opportunity). At a minimum, we would like to have a process for keeping track of educational events that feature MNE-Python or other tools in the MNE suite. Ideally, we would go further and initiate a recurring series of tutorials that could be advertised widely. Such events might even provide a small revenue stream for MNE-Python, to support things like continuous integration costs.
In progress#
Diversity, Equity, and Inclusion (DEI)#
MNE-Python is committed to recruiting and retaining a diverse pool of contributors, see #8221.
First-class OPM support#
MNE-Python has support for reading some OPM data formats such as FIF and FIL/QuSpin. Support should be added for other manufacturers, and standard preprocessing routines should be added to deal with coregistration adjustment and OPM-specific artifacts. See for example #11275, #11276, #11579, #12179.
Deep source modeling#
Existing source modeling and inverse routines are not explicitly designed to deal with deep sources. Advanced algorithms exist from MGH for enhancing deep source localization, and these should be implemented and vetted in MNE-Python. See #6784.
Time-frequency classes#
Historically our codebase had classes related to TFRs that were incomplete. New classes are being built from the ground up:
new classes
SpectrumandEpochsSpectrum(implemented in #10184, with follow-up tweaks and bugfixes in #11178, #11259, #11280, #11345, #11418, #11563, #11680, #11682, #11778, #11921, #11978, #12747), and corresponding array-based constructorsSpectrumArrayandEpochsSpectrumArray(#11803).new class
RawTFRand updated classesEpochsTFRandAverageTFR, and corresponding array-based constructorsRawTFRArray,EpochsTFRArrayandAverageTFRArray(implemented in #11282, with follow-ups in #12514, #12842).new/updated classes for source-space frequency and time-frequency data are not yet implemented.
Other related issues: #6290, #7671, #8026, #8724, #9045, and PRs: #6609, #6629, #6672, #6673, #8397, #8892.
Modernization of realtime processing#
LSL has become the de facto standard for streaming data from EEG/MEG systems. We should deprecate MNE-Realtime in favor of the newly minted MNE-LSL. We should then fully support MNE-LSL using modern coding best practices such as CI integration.
Core components of commonly used real-time processing pipelines should be implemented in MNE-LSL, including but not limited to realtime IIR filtering, artifact rejection, montage and reference setting, and online averaging. Integration with standard MNE-Python plotting routines (evoked joint plots, topomaps, etc.) should be supported with continuous updating.
Clustering statistics API#
The current clustering statistics code has limited functionality. It should be
re-worked to create a new cluster_based_statistic or similar function.
The new API will likely be along the lines of:
cluster_stat(obs, design, *, alpha=0.05, cluster_alpha=0.05, ...)
with:
obspandas.DataFrameHas columns like “subject”, “condition”, and “data”. The “data” column holds things like
mne.Evoked,mne.SourceEstimate,mne.time_frequency.Spectrum, etc.designstrLikely Wilkinson notation to mirror
patsy.dmatrices()(e.g., this is is used bystatsmodels.regression.linear_model.OLS). Getting from the string to the design matrix could be done via Patsy or more likely Formulaic.
This generic API will support mixed within- and between-subjects designs, different statistical functions/tests, etc. This should be achievable without introducing any significant speed penalty (e.g., < 10% slower) compared to the existing more specialized/limited functions, since most computation cost is in clustering rather than statistical testing.
The clustering function will return a user-friendly ClusterStat object or similar
that retains information about dimensionality, significance, etc. and facilitates
plotting and interpretation of results.
Clear tutorials will be needed to:
Show how different contrasts can be done (toy data).
Show some common analyses on real data (time-freq, sensor space, source space, etc.)
Regression tests will be written to ensure equivalent outputs when compared to FieldTrip for cases that FieldTrip also supports.
Documentation updates#
Our documentation has many minor issues, which can be found under the tag #labels/DOC.
Completed#
3D visualization#
Historically we used Mayavi for 3D visualization, but faced limitations and challenges with it. We switched to PyVista to get major improvements, such as:
Proper notebook support (through ``ipyvtklink``) (complete; updated to use
trame)Better interactivity with surface plots (complete)
Time-frequency plotting (complementary to volume-based Time-frequency visualization)
Integration of multiple functions as done in
mne_analyze, e.g., simultaneous source estimate viewing, field map viewing, head surface display, etc. These were all available in separate functions, but can now be combined in a single plot.
The meta-issue tracking to-do lists for surface plotting was #7162.
Improved sEEG/ECoG/DBS support#
iEEG-specific pipeline steps such as electrode localization and visualizations are now available in MNE-gui-addons.
Access to open EEG/MEG databases#
Open EEG/MEG databases are now more easily accessible via standardized tools such as openneuro-py.
Eye-tracking support#
We had a GSoC student funded to improve support for eye-tracking data, see the GSoC proposal for details. An EyeLink data reader and analysis/plotting functions are now available.
Pediatric and clinical MEG pipelines#
MNE-Python provides automated analysis of BIDS-compliant datasets via MNE-BIDS-Pipeline. Functionality from the mnefun pipeline, which has been used extensively for pediatric data analysis at I-LABS, now provides better support for pediatric and clinical data processing. Multiple processing steps (e.g., eSSS), sanity checks (e.g., cHPI quality), and reporting (e.g., SSP joint plots, SNR plots) have been added.
Integrate OpenMEEG via improved Python bindings#
OpenMEEG is a state-of-the art solver for forward modeling in the field of brain imaging with MEG/EEG. It solves numerically partial differential equations (PDE). It is written in C++ with Python bindings written in SWIG. The ambition of the project is to integrate OpenMEEG into MNE offering to MNE the ability to solve more forward problems (cortical mapping, intracranial recordings, etc.). Tasks that have been completed:
Cleanup Python bindings (remove useless functions, check memory managements, etc.)
Understand how MNE encodes info about sensors (location, orientation, integration points etc.) and allow OpenMEEG to be used.
Modernize CI systems (e.g., using
cibuildwheel).Automated deployment on PyPI and conda-forge.
Time-frequency visualization#
We implemented a viewer for interactive visualization of volumetric source-time-frequency (5-D) maps on MRI slices (orthogonal 2D viewer). NutmegTrip (written by Sarang Dalal) provides similar functionality in MATLAB in conjunction with FieldTrip. Example of NutmegTrip’s source-time-frequency mode in action (click for link to YouTube):
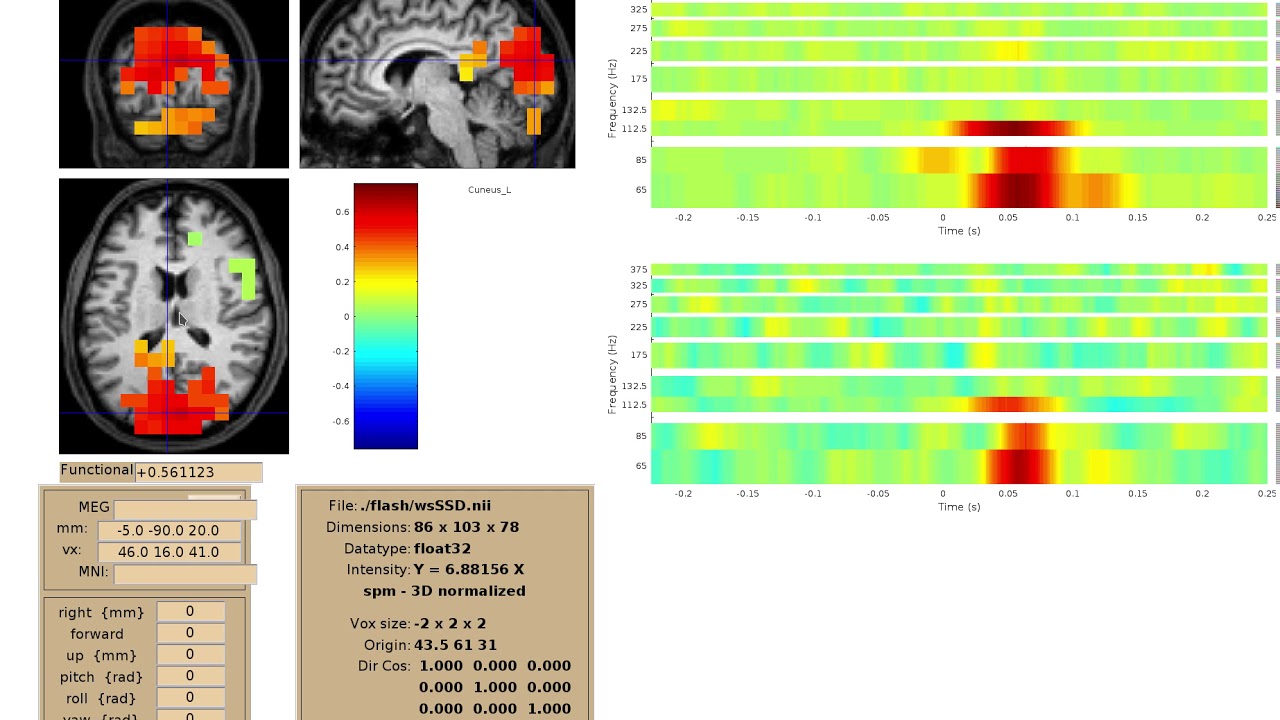
Distributed computing support#
MNE-BIDS-Pipeline has been enhanced with support for cloud computing via Dask and joblib. After configuring Dask to use local or remote distributed computing resources, MNE-BIDS-Pipeline can readily make use of remote workers to parallelize processing across subjects.
2D visualization#
This goal was completed under CZI EOSS2. Some additional enhancements that could also be implemented are listed in #7751.
Tutorial / example overhaul#
This goal was completed under CZI EOSS2. Ongoing documentation needs are listed in Documentation updates.
Cluster computing images#
As part of this goal, we created docker images suitable for cloud computing via MNE-Docker.