Note
Go to the end to download the full example code
Interpolate bad channels for MEG/EEG channels#
This example shows how to interpolate bad MEG/EEG channels
Using spherical splines from [1] for EEG data.
Using field interpolation for MEG and EEG data.
In this example, the bad channels will still be marked as bad. Only the data in those channels is replaced.
# Authors: Denis A. Engemann <denis.engemann@gmail.com>
# Mainak Jas <mainak.jas@telecom-paristech.fr>
#
# License: BSD-3-Clause
import mne
from mne.datasets import sample
print(__doc__)
data_path = sample.data_path()
meg_path = data_path / 'MEG' / 'sample'
fname = meg_path / 'sample_audvis-ave.fif'
evoked = mne.read_evokeds(fname, condition='Left Auditory',
baseline=(None, 0))
# plot with bads
evoked.plot(exclude=[], picks=('grad', 'eeg'))
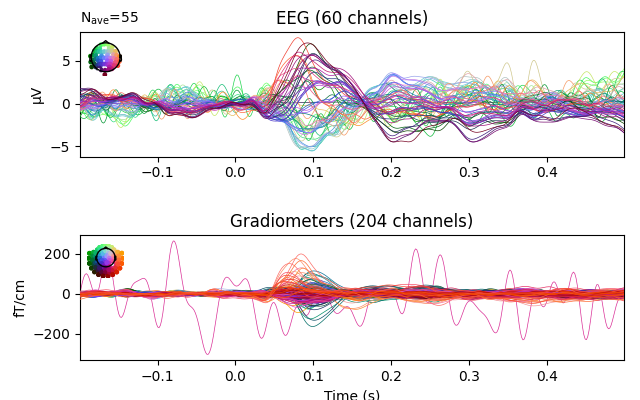
Reading /home/circleci/mne_data/MNE-sample-data/MEG/sample/sample_audvis-ave.fif ...
Read a total of 4 projection items:
PCA-v1 (1 x 102) active
PCA-v2 (1 x 102) active
PCA-v3 (1 x 102) active
Average EEG reference (1 x 60) active
Found the data of interest:
t = -199.80 ... 499.49 ms (Left Auditory)
0 CTF compensation matrices available
nave = 55 - aspect type = 100
Projections have already been applied. Setting proj attribute to True.
Applying baseline correction (mode: mean)
Compute interpolation (also works with Raw and Epochs objects)
evoked_interp = evoked.copy().interpolate_bads(reset_bads=False)
evoked_interp.plot(exclude=[], picks=('grad', 'eeg'))
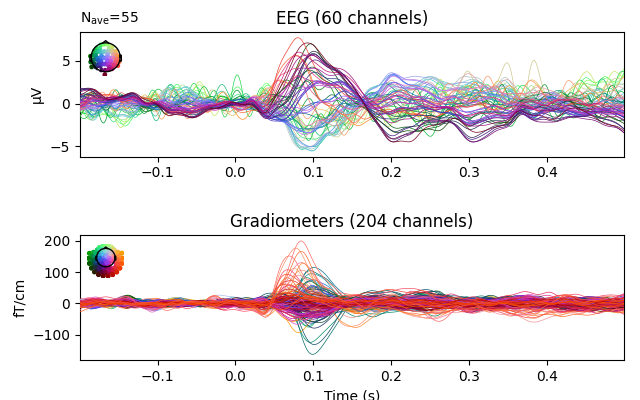
Interpolating bad channels
Automatic origin fit: head of radius 91.0 mm
Computing interpolation matrix from 59 sensor positions
Interpolating 1 sensors
Computing dot products for 305 MEG channels...
[Parallel(n_jobs=1)]: Using backend SequentialBackend with 1 concurrent workers.
[Parallel(n_jobs=1)]: Done 1 out of 1 | elapsed: 4.6s remaining: 0.0s
[Parallel(n_jobs=1)]: Done 1 out of 1 | elapsed: 4.6s finished
Computing cross products for 305 → 1 MEG channel...
Preparing the mapping matrix...
Truncating at 85/305 components to omit less than 0.0001 (9.4e-05)
You can also use minimum-norm for EEG as well as MEG
evoked_interp_mne = evoked.copy().interpolate_bads(
reset_bads=False, method=dict(eeg='MNE'), verbose=True)
evoked_interp_mne.plot(exclude=[], picks=('grad', 'eeg'))
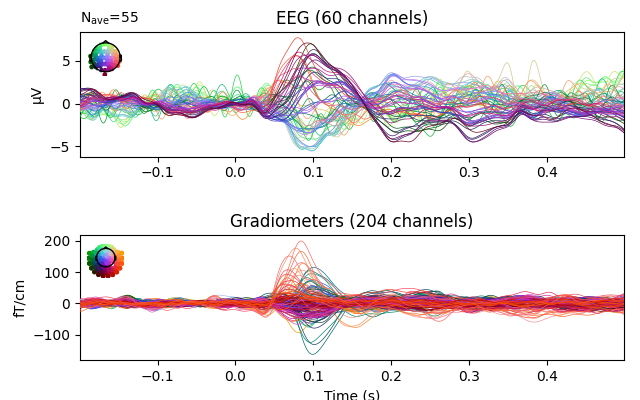
Interpolating bad channels
Automatic origin fit: head of radius 91.0 mm
Computing dot products for 305 MEG channels...
[Parallel(n_jobs=1)]: Using backend SequentialBackend with 1 concurrent workers.
[Parallel(n_jobs=1)]: Done 1 out of 1 | elapsed: 4.7s remaining: 0.0s
[Parallel(n_jobs=1)]: Done 1 out of 1 | elapsed: 4.7s finished
Computing cross products for 305 → 1 MEG channel...
Preparing the mapping matrix...
Truncating at 85/305 components to omit less than 0.0001 (9.4e-05)
Computing dot products for 59 EEG channels...
[Parallel(n_jobs=1)]: Using backend SequentialBackend with 1 concurrent workers.
[Parallel(n_jobs=1)]: Done 1 out of 1 | elapsed: 0.1s remaining: 0.0s
[Parallel(n_jobs=1)]: Done 1 out of 1 | elapsed: 0.1s finished
Computing cross products for 59 → 60 EEG channels...
Preparing the mapping matrix...
Truncating at 58/59 components and regularizing with α=1.0e-01
The map has an average electrode reference (60 channels)
References#
Total running time of the script: ( 0 minutes 15.258 seconds)
Estimated memory usage: 12 MB