Note
Go to the end to download the full example code.
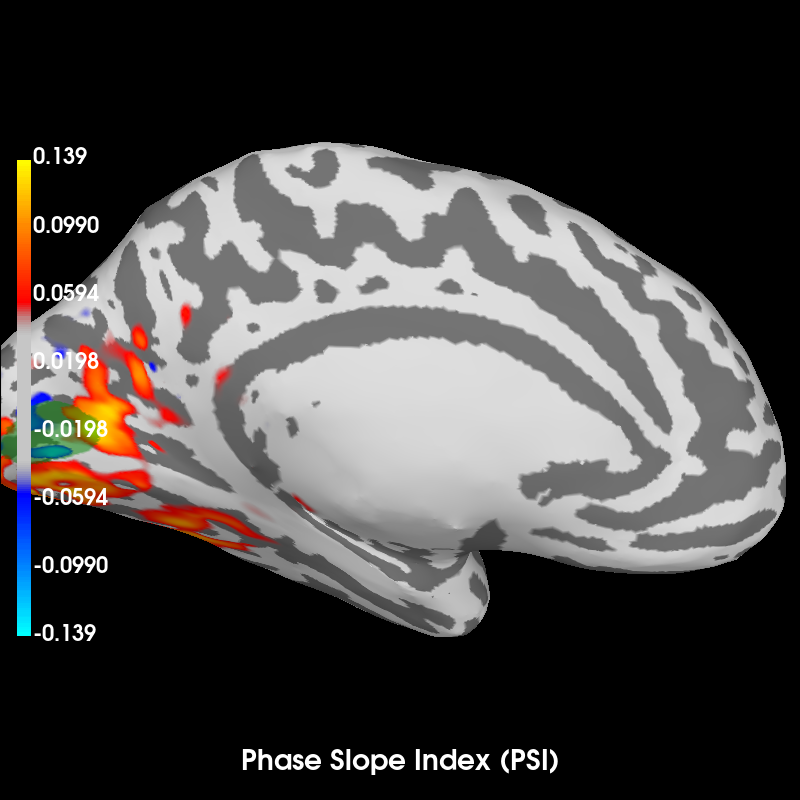
Compute Phase Slope Index (PSI) in source space for a visual stimulus#
This example demonstrates how the phase slope index (PSI) [1] can be computed in source space based on single trial dSPM source estimates. In addition, the example shows advanced usage of the connectivity estimation routines by first extracting a label time course for each epoch and then combining the label time course with the single trial source estimates to compute the connectivity.
The result clearly shows how the activity in the visual label precedes more widespread activity (as a postivive PSI means the label time course is leading).
# Author: Martin Luessi <mluessi@nmr.mgh.harvard.edu>
#
# License: BSD (3-clause)
import mne
import numpy as np
from mne.datasets import sample
from mne.minimum_norm import apply_inverse_epochs, read_inverse_operator
from mne_connectivity import phase_slope_index, seed_target_indices
print(__doc__)
data_path = sample.data_path()
subjects_dir = data_path / "subjects"
fname_inv = data_path / "MEG/sample/sample_audvis-meg-oct-6-meg-inv.fif"
fname_raw = data_path / "MEG/sample/sample_audvis_filt-0-40_raw.fif"
fname_event = data_path / "MEG/sample/sample_audvis_filt-0-40_raw-eve.fif"
fname_label = data_path / "MEG/sample/labels/Vis-lh.label"
event_id, tmin, tmax = 4, -0.2, 0.5
method = "dSPM" # use dSPM method (could also be MNE or sLORETA)
# Load data
inverse_operator = read_inverse_operator(fname_inv)
raw = mne.io.read_raw_fif(fname_raw, preload=True)
events = mne.read_events(fname_event)
# pick MEG channels
picks = mne.pick_types(
raw.info, meg=True, eeg=False, stim=False, eog=True, exclude="bads"
)
# Read epochs
epochs = mne.Epochs(
raw,
events,
event_id,
tmin,
tmax,
picks=picks,
baseline=(None, 0),
reject=dict(mag=4e-12, grad=4000e-13, eog=150e-6),
)
# Compute inverse solution and for each epoch. Note that since we are passing
# the output to both extract_label_time_course and the phase_slope_index
# functions, we have to use "return_generator=False", since it is only possible
# to iterate over generators once.
snr = 1.0 # use lower SNR for single epochs
lambda2 = 1.0 / snr**2
stcs = apply_inverse_epochs(
epochs, inverse_operator, lambda2, method, pick_ori="normal", return_generator=True
)
# Now, we generate seed time series by averaging the activity in the left
# visual corex
label = mne.read_label(fname_label)
src = inverse_operator["src"] # the source space used
seed_ts = mne.extract_label_time_course(
stcs, label, src, mode="mean_flip", verbose="error"
)
# Combine the seed time course with the source estimates. There will be a total
# of 7500 signals:
# index 0: time course extracted from label
# index 1..7499: dSPM source space time courses
stcs = apply_inverse_epochs(
epochs, inverse_operator, lambda2, method, pick_ori="normal", return_generator=True
)
comb_ts = list(zip(seed_ts, stcs))
# Construct indices to estimate connectivity between the label time course
# and all source space time courses
vertices = [src[i]["vertno"] for i in range(2)]
n_signals_tot = 1 + len(vertices[0]) + len(vertices[1])
indices = seed_target_indices([0], np.arange(1, n_signals_tot))
# Compute the PSI in the frequency range 10Hz-20Hz. We exclude the baseline
# period from the connectivity estimation.
fmin = 10.0
fmax = 20.0
tmin_con = 0.0
sfreq = epochs.info["sfreq"] # the sampling frequency
psi = phase_slope_index(
comb_ts,
mode="multitaper",
indices=indices,
sfreq=sfreq,
fmin=fmin,
fmax=fmax,
tmin=tmin_con,
)
# Generate a SourceEstimate with the PSI. This is simple since we used a single
# seed (inspect the indices variable to see how the PSI scores are arranged in
# the output)
psi_stc = mne.SourceEstimate(
psi.get_data(), vertices=vertices, tmin=0, tstep=1, subject="sample"
)
# Now we can visualize the PSI using the :meth:`~mne.SourceEstimate.plot`
# method. We use a custom colormap to show signed values
v_max = np.max(np.abs(psi.get_data()))
brain = psi_stc.plot(
surface="inflated",
hemi="lh",
time_label="Phase Slope Index (PSI)",
subjects_dir=subjects_dir,
clim=dict(kind="percent", pos_lims=(95, 97.5, 100)),
)
brain.show_view("medial")
brain.add_label(str(fname_label), color="green", alpha=0.7)
Reading inverse operator decomposition from /home/circleci/mne_data/MNE-sample-data/MEG/sample/sample_audvis-meg-oct-6-meg-inv.fif...
Reading inverse operator info...
[done]
Reading inverse operator decomposition...
[done]
305 x 305 full covariance (kind = 1) found.
Read a total of 4 projection items:
PCA-v1 (1 x 102) active
PCA-v2 (1 x 102) active
PCA-v3 (1 x 102) active
Average EEG reference (1 x 60) active
Noise covariance matrix read.
22494 x 22494 diagonal covariance (kind = 2) found.
Source covariance matrix read.
22494 x 22494 diagonal covariance (kind = 6) found.
Orientation priors read.
22494 x 22494 diagonal covariance (kind = 5) found.
Depth priors read.
Did not find the desired covariance matrix (kind = 3)
Reading a source space...
Computing patch statistics...
Patch information added...
Distance information added...
[done]
Reading a source space...
Computing patch statistics...
Patch information added...
Distance information added...
[done]
2 source spaces read
Read a total of 4 projection items:
PCA-v1 (1 x 102) active
PCA-v2 (1 x 102) active
PCA-v3 (1 x 102) active
Average EEG reference (1 x 60) active
Source spaces transformed to the inverse solution coordinate frame
Opening raw data file /home/circleci/mne_data/MNE-sample-data/MEG/sample/sample_audvis_filt-0-40_raw.fif...
Read a total of 4 projection items:
PCA-v1 (1 x 102) idle
PCA-v2 (1 x 102) idle
PCA-v3 (1 x 102) idle
Average EEG reference (1 x 60) idle
Range : 6450 ... 48149 = 42.956 ... 320.665 secs
Ready.
Reading 0 ... 41699 = 0.000 ... 277.709 secs...
Not setting metadata
70 matching events found
Setting baseline interval to [-0.19979521315838786, 0.0] s
Applying baseline correction (mode: mean)
Created an SSP operator (subspace dimension = 3)
3 projection items activated
Preparing the inverse operator for use...
Scaled noise and source covariance from nave = 1 to nave = 1
Created the regularized inverter
Created an SSP operator (subspace dimension = 3)
Created the whitener using a noise covariance matrix with rank 302 (3 small eigenvalues omitted)
Computing noise-normalization factors (dSPM)...
[done]
Picked 305 channels from the data
Computing inverse...
Eigenleads need to be weighted ...
Processing epoch : 1 / 70 (at most)
Processing epoch : 2 / 70 (at most)
Processing epoch : 3 / 70 (at most)
Processing epoch : 4 / 70 (at most)
Processing epoch : 5 / 70 (at most)
Processing epoch : 6 / 70 (at most)
Processing epoch : 7 / 70 (at most)
Rejecting epoch based on EOG : ['EOG 061']
Processing epoch : 8 / 70 (at most)
Processing epoch : 9 / 70 (at most)
Processing epoch : 10 / 70 (at most)
Processing epoch : 11 / 70 (at most)
Rejecting epoch based on EOG : ['EOG 061']
Processing epoch : 12 / 70 (at most)
Processing epoch : 13 / 70 (at most)
Processing epoch : 14 / 70 (at most)
Processing epoch : 15 / 70 (at most)
Rejecting epoch based on EOG : ['EOG 061']
Processing epoch : 16 / 70 (at most)
Processing epoch : 17 / 70 (at most)
Processing epoch : 18 / 70 (at most)
Processing epoch : 19 / 70 (at most)
Rejecting epoch based on EOG : ['EOG 061']
Processing epoch : 20 / 70 (at most)
Processing epoch : 21 / 70 (at most)
Processing epoch : 22 / 70 (at most)
Processing epoch : 23 / 70 (at most)
Processing epoch : 24 / 70 (at most)
Processing epoch : 25 / 70 (at most)
Rejecting epoch based on EOG : ['EOG 061']
Processing epoch : 26 / 70 (at most)
Processing epoch : 27 / 70 (at most)
Rejecting epoch based on EOG : ['EOG 061']
Processing epoch : 28 / 70 (at most)
Processing epoch : 29 / 70 (at most)
Processing epoch : 30 / 70 (at most)
Processing epoch : 31 / 70 (at most)
Processing epoch : 32 / 70 (at most)
Processing epoch : 33 / 70 (at most)
Processing epoch : 34 / 70 (at most)
Rejecting epoch based on EOG : ['EOG 061']
Processing epoch : 35 / 70 (at most)
Processing epoch : 36 / 70 (at most)
Processing epoch : 37 / 70 (at most)
Processing epoch : 38 / 70 (at most)
Rejecting epoch based on EOG : ['EOG 061']
Processing epoch : 39 / 70 (at most)
Rejecting epoch based on EOG : ['EOG 061']
Processing epoch : 40 / 70 (at most)
Processing epoch : 41 / 70 (at most)
Processing epoch : 42 / 70 (at most)
Processing epoch : 43 / 70 (at most)
Processing epoch : 44 / 70 (at most)
Processing epoch : 45 / 70 (at most)
Rejecting epoch based on EOG : ['EOG 061']
Processing epoch : 46 / 70 (at most)
Processing epoch : 47 / 70 (at most)
Processing epoch : 48 / 70 (at most)
Processing epoch : 49 / 70 (at most)
Processing epoch : 50 / 70 (at most)
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Processing epoch : 51 / 70 (at most)
Processing epoch : 52 / 70 (at most)
Processing epoch : 53 / 70 (at most)
Processing epoch : 54 / 70 (at most)
Processing epoch : 55 / 70 (at most)
Rejecting epoch based on EOG : ['EOG 061']
Processing epoch : 56 / 70 (at most)
Estimating phase slope index (PSI)
Connectivity computation...
computing connectivity for 7498 connections
using t=0.000s..0.499s for estimation (76 points)
frequencies: 11.9Hz..19.8Hz (5 points)
Using multitaper spectrum estimation with 7 DPSS windows
the following metrics will be computed: Coherency
computing cross-spectral density for epoch 1
computing cross-spectral density for epoch 2
computing cross-spectral density for epoch 3
computing cross-spectral density for epoch 4
computing cross-spectral density for epoch 5
computing cross-spectral density for epoch 6
computing cross-spectral density for epoch 7
computing cross-spectral density for epoch 8
computing cross-spectral density for epoch 9
computing cross-spectral density for epoch 10
computing cross-spectral density for epoch 11
computing cross-spectral density for epoch 12
computing cross-spectral density for epoch 13
computing cross-spectral density for epoch 14
computing cross-spectral density for epoch 15
computing cross-spectral density for epoch 16
computing cross-spectral density for epoch 17
computing cross-spectral density for epoch 18
computing cross-spectral density for epoch 19
computing cross-spectral density for epoch 20
computing cross-spectral density for epoch 21
computing cross-spectral density for epoch 22
computing cross-spectral density for epoch 23
computing cross-spectral density for epoch 24
computing cross-spectral density for epoch 25
computing cross-spectral density for epoch 26
computing cross-spectral density for epoch 27
computing cross-spectral density for epoch 28
computing cross-spectral density for epoch 29
computing cross-spectral density for epoch 30
computing cross-spectral density for epoch 31
computing cross-spectral density for epoch 32
computing cross-spectral density for epoch 33
computing cross-spectral density for epoch 34
computing cross-spectral density for epoch 35
computing cross-spectral density for epoch 36
computing cross-spectral density for epoch 37
computing cross-spectral density for epoch 38
computing cross-spectral density for epoch 39
computing cross-spectral density for epoch 40
computing cross-spectral density for epoch 41
computing cross-spectral density for epoch 42
computing cross-spectral density for epoch 43
computing cross-spectral density for epoch 44
computing cross-spectral density for epoch 45
computing cross-spectral density for epoch 46
computing cross-spectral density for epoch 47
computing cross-spectral density for epoch 48
computing cross-spectral density for epoch 49
computing cross-spectral density for epoch 50
computing cross-spectral density for epoch 51
computing cross-spectral density for epoch 52
computing cross-spectral density for epoch 53
computing cross-spectral density for epoch 54
computing cross-spectral density for epoch 55
computing cross-spectral density for epoch 56
[Connectivity computation done]
Computing PSI from estimated Coherency: <SpectralConnectivity | freq : [11.854243, 19.757072], , nave : 56, nodes, n_estimated : 7499, 7498, ~1.2 MB>
[PSI Estimation Done]
Using control points [0.03622925 0.05660114 0.13862278]
References#
Total running time of the script: (0 minutes 14.998 seconds)