Note
Go to the end to download the full example code.
Compute seed-based time-frequency connectivity in sensor space#
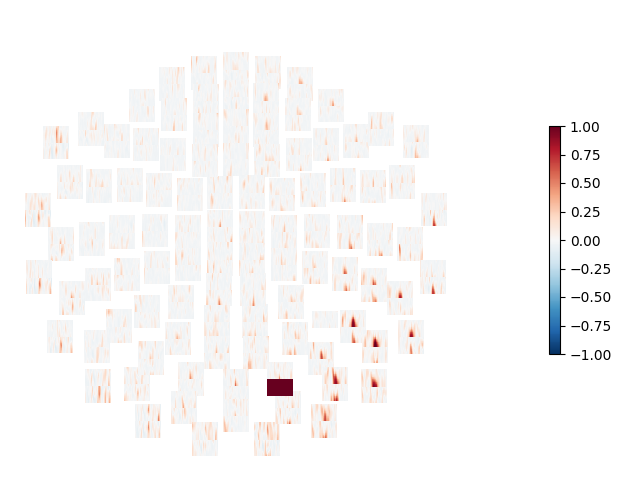
Computes the connectivity between a seed-gradiometer close to the visual cortex and all other gradiometers. The connectivity is computed in the time-frequency domain using Morlet wavelets and the debiased squared weighted phase lag index [1] is used as connectivity metric.
# Author: Martin Luessi <mluessi@nmr.mgh.harvard.edu>
#
# License: BSD (3-clause)
import os.path as op
import mne
import numpy as np
from mne import io
from mne.datasets import sample
from mne.time_frequency import AverageTFRArray
from mne_connectivity import seed_target_indices, spectral_connectivity_epochs
print(__doc__)
Set parameters
data_path = sample.data_path()
raw_fname = op.join(data_path, "MEG", "sample", "sample_audvis_filt-0-40_raw.fif")
event_fname = op.join(data_path, "MEG", "sample", "sample_audvis_filt-0-40_raw-eve.fif")
# Setup for reading the raw data
raw = io.read_raw_fif(raw_fname)
events = mne.read_events(event_fname)
# Add a bad channel
raw.info["bads"] += ["MEG 2443"]
# Pick MEG gradiometers
picks = mne.pick_types(
raw.info, meg="grad", eeg=False, stim=False, eog=True, exclude="bads"
)
# Create epochs for left-visual condition
event_id, tmin, tmax = 3, -0.2, 0.5
epochs = mne.Epochs(
raw,
events,
event_id,
tmin,
tmax,
picks=picks,
baseline=(None, 0),
reject=dict(grad=4000e-13, eog=150e-6),
preload=True,
)
# Use 'MEG 2343' as seed
seed_ch = "MEG 2343"
picks_ch_names = [raw.ch_names[i] for i in picks]
# Create seed-target indices for connectivity computation
seed = picks_ch_names.index(seed_ch)
targets = np.arange(len(picks))
indices = seed_target_indices(seed, targets)
# Define wavelet frequencies and number of cycles
cwt_freqs = np.arange(7, 30, 2)
cwt_n_cycles = cwt_freqs / 7.0
# Run the connectivity analysis using 2 parallel jobs
sfreq = raw.info["sfreq"] # the sampling frequency
con = spectral_connectivity_epochs(
epochs,
indices=indices,
method="wpli2_debiased",
mode="cwt_morlet",
sfreq=sfreq,
cwt_freqs=cwt_freqs,
cwt_n_cycles=cwt_n_cycles,
n_jobs=1,
)
times = con.times
freqs = con.freqs
# Mark the seed channel with a value of 1.0, so we can see it in the plot
con.get_data()[np.where(indices[1] == seed)] = 1.0
# Show topography of connectivity from seed
title = "WPLI2 - Visual - Seed %s" % seed_ch
layout = mne.find_layout(epochs.info, "meg") # use full layout
# Note that users of mne < 1.7 should use the `AverageTFR` class
tfr = AverageTFRArray(epochs.info, con.get_data(), times, freqs, nave=len(epochs))
tfr.plot_topo(fig_facecolor="w", font_color="k", border="k")
Opening raw data file /home/circleci/mne_data/MNE-sample-data/MEG/sample/sample_audvis_filt-0-40_raw.fif...
Read a total of 4 projection items:
PCA-v1 (1 x 102) idle
PCA-v2 (1 x 102) idle
PCA-v3 (1 x 102) idle
Average EEG reference (1 x 60) idle
Range : 6450 ... 48149 = 42.956 ... 320.665 secs
Ready.
Not setting metadata
73 matching events found
Setting baseline interval to [-0.19979521315838786, 0.0] s
Applying baseline correction (mode: mean)
0 projection items activated
Loading data for 73 events and 106 original time points ...
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
6 bad epochs dropped
Adding metadata with 3 columns
Connectivity computation...
computing connectivity for 204 connections
using t=-0.200s..0.499s for estimation (106 points)
frequencies: 9.0Hz..29.0Hz (11 points)
using CWT with Morlet wavelets to estimate spectra
the following metrics will be computed: Debiased WPLI Square
computing cross-spectral density for epoch 1
computing cross-spectral density for epoch 2
computing cross-spectral density for epoch 3
computing cross-spectral density for epoch 4
computing cross-spectral density for epoch 5
computing cross-spectral density for epoch 6
computing cross-spectral density for epoch 7
computing cross-spectral density for epoch 8
computing cross-spectral density for epoch 9
computing cross-spectral density for epoch 10
computing cross-spectral density for epoch 11
computing cross-spectral density for epoch 12
computing cross-spectral density for epoch 13
computing cross-spectral density for epoch 14
computing cross-spectral density for epoch 15
computing cross-spectral density for epoch 16
computing cross-spectral density for epoch 17
computing cross-spectral density for epoch 18
computing cross-spectral density for epoch 19
computing cross-spectral density for epoch 20
computing cross-spectral density for epoch 21
computing cross-spectral density for epoch 22
computing cross-spectral density for epoch 23
computing cross-spectral density for epoch 24
computing cross-spectral density for epoch 25
computing cross-spectral density for epoch 26
computing cross-spectral density for epoch 27
computing cross-spectral density for epoch 28
computing cross-spectral density for epoch 29
computing cross-spectral density for epoch 30
computing cross-spectral density for epoch 31
computing cross-spectral density for epoch 32
computing cross-spectral density for epoch 33
computing cross-spectral density for epoch 34
computing cross-spectral density for epoch 35
computing cross-spectral density for epoch 36
computing cross-spectral density for epoch 37
computing cross-spectral density for epoch 38
computing cross-spectral density for epoch 39
computing cross-spectral density for epoch 40
computing cross-spectral density for epoch 41
computing cross-spectral density for epoch 42
computing cross-spectral density for epoch 43
computing cross-spectral density for epoch 44
computing cross-spectral density for epoch 45
computing cross-spectral density for epoch 46
computing cross-spectral density for epoch 47
computing cross-spectral density for epoch 48
computing cross-spectral density for epoch 49
computing cross-spectral density for epoch 50
computing cross-spectral density for epoch 51
computing cross-spectral density for epoch 52
computing cross-spectral density for epoch 53
computing cross-spectral density for epoch 54
computing cross-spectral density for epoch 55
computing cross-spectral density for epoch 56
computing cross-spectral density for epoch 57
computing cross-spectral density for epoch 58
computing cross-spectral density for epoch 59
computing cross-spectral density for epoch 60
computing cross-spectral density for epoch 61
computing cross-spectral density for epoch 62
computing cross-spectral density for epoch 63
computing cross-spectral density for epoch 64
computing cross-spectral density for epoch 65
computing cross-spectral density for epoch 66
computing cross-spectral density for epoch 67
[Connectivity computation done]
No baseline correction applied
<Figure size 640x480 with 2 Axes>
References#
Total running time of the script: (0 minutes 3.978 seconds)