Note
Go to the end to download the full example code.
Volume Source Time Course Estimate for a Group Study#
In this example, we’ll show how to use the MNE volume stc viewing GUI to compare brain activity across subjects in a group study.
# Authors: Alex Rockhill <aprockhill@mailbox.org>
#
# License: BSD-3-Clause
import os.path as op
import autoreject
import mne
import numpy as np
from mne.time_frequency import csd_tfr
import mne_gui_addons as mne_gui
fs_dir = mne.datasets.fetch_fsaverage(verbose=True)
subjects_dir = op.dirname(fs_dir)
template = "fsaverage"
trans = "fsaverage" # MNE has a built-in fsaverage transformation
src = mne.read_source_spaces(op.join(fs_dir, "bem", "fsaverage-vol-5-src.fif"))
bem = mne.read_bem_solution(
op.join(fs_dir, "bem", "fsaverage-5120-5120-5120-bem-sol.fif")
)
# basic task parameters
tmin, tmax = -1.0, 4.0
active_win = (0, 4)
baseline_win = (-1, 0)
event_id = dict(hands=2, feet=3)
runs = [6, 10, 14] # motor imagery: hands vs feet
0 files missing from root.txt in /home/circleci/mne_data/MNE-sample-data/subjects
0 files missing from bem.txt in /home/circleci/mne_data/MNE-sample-data/subjects/fsaverage
Reading a source space...
[done]
1 source spaces read
Loading surfaces...
Loading the solution matrix...
Three-layer model surfaces loaded.
Loaded linear collocation BEM solution from /home/circleci/mne_data/MNE-sample-data/subjects/fsaverage/bem/fsaverage-5120-5120-5120-bem-sol.fif
Compute source time course (stc) estimates for both time courses and spectrograms (time-frequency).
montage = mne.channels.make_standard_montage("standard_1005")
stcs_tfr = list()
stcs_epochs = list()
insts_tfr = list()
insts_epochs = list()
for sub in range(1, 4):
print(f"Computing source estimate for subject {sub}")
raw_fnames = mne.datasets.eegbci.load_data(subjects=[sub], runs=runs, update_path=True)
raw = mne.concatenate_raws(
[
mne.io.read_raw(raw_fname, preload=True, verbose=False)
for raw_fname in raw_fnames
]
)
mne.datasets.eegbci.standardize(raw) # set channel names
raw.set_montage(montage, verbose=False)
# make epochs
events, _ = mne.events_from_annotations(raw, event_id=dict(T1=2, T2=3))
picks = mne.pick_types(
raw.info, meg=False, eeg=True, stim=False, eog=False, exclude="bads"
)
epochs = mne.Epochs(
raw,
events,
event_id,
tmin - 0.5,
tmax + 0.5,
proj=True,
picks=picks,
baseline=None,
preload=True,
)
del raw
epochs.set_eeg_reference(projection=True)
# reject bad epochs
reject = autoreject.get_rejection_threshold(epochs)
epochs.drop_bad(reject=reject)
epochs.filter(l_freq=1, h_freq=None)
ica = mne.preprocessing.ICA(n_components=15, random_state=11)
ica.fit(epochs)
eog_idx, scores = ica.find_bads_eog(epochs, ch_name="Fp1")
muscle_idx, scores = ica.find_bads_muscle(epochs)
ica.apply(epochs, exclude=eog_idx + muscle_idx)
# make forward model
fwd = mne.make_forward_solution(
epochs.info, trans=trans, src=src, bem=bem, eeg=True, mindist=5.0
)
rank = mne.compute_rank(epochs, tol=1e-6, tol_kind="relative")
# compute cross-spectral density matrices
freqs = np.logspace(np.log10(12), np.log10(30), 9)
# time-frequency decomposition
epochs_tfr = mne.time_frequency.tfr_morlet(
epochs,
freqs=freqs,
n_cycles=freqs / 2,
return_itc=False,
average=False,
output="complex",
)
baseline_csd = csd_tfr(epochs_tfr, tmin=baseline_win[0], tmax=baseline_win[1])
epochs_tfr.decimate(20) # decimate for speed
insts_tfr.append(epochs_tfr)
# Compute source estimate using MNE solver
snr = 3.0
lambda2 = 1.0 / snr**2
method = "MNE" # use MNE method (could also be dSPM or sLORETA)
epochs.decimate(20)
insts_epochs.append(epochs)
# do time-series epochs first
baseline_cov = mne.compute_covariance(epochs, tmax=0)
inverse_operator = mne.minimum_norm.make_inverse_operator(
epochs.info, fwd, baseline_cov
)
stcs_epochs.append(
mne.minimum_norm.apply_inverse_epochs(
epochs,
inverse_operator,
lambda2,
method=method,
pick_ori="vector",
return_generator=True,
)
)
# make a different inverse operator for each frequency so as to properly
# whiten the sensor data
stcs = list()
for freq_idx in range(epochs_tfr.freqs.size):
# for each frequency, compute a separate covariance matrix
baseline_cov = baseline_csd.get_data(index=freq_idx, as_cov=True)
# only normalize by real
baseline_cov["data"] = baseline_cov["data"].real
# then use that covariance matrix as normalization for the inverse
# operator
inverse_operator = mne.minimum_norm.make_inverse_operator(
epochs.info, fwd, baseline_cov
)
# finally, compute the stcs for each epoch and frequency
stcs = mne.minimum_norm.apply_inverse_tfr_epochs(
epochs_tfr,
inverse_operator,
lambda2,
method=method,
pick_ori="vector",
return_generator=True,
)
# append to group
stcs_tfr.append(stcs)
Computing source estimate for subject 1
Using default location ~/mne_data for EEGBCI...
Downloading EEGBCI data
Download complete in 19s (7.4 MB)
Used Annotations descriptions: [np.str_('T1'), np.str_('T2')]
Not setting metadata
45 matching events found
No baseline correction applied
0 projection items activated
Using data from preloaded Raw for 45 events and 961 original time points ...
0 bad epochs dropped
EEG channel type selected for re-referencing
Adding average EEG reference projection.
1 projection items deactivated
Average reference projection was added, but has not been applied yet. Use the apply_proj method to apply it.
Estimating rejection dictionary for eeg
Rejecting epoch based on EEG : ['Fp1']
Rejecting epoch based on EEG : ['Fp1', 'AF7']
Rejecting epoch based on EEG : ['FC5', 'FC3', 'FC1', 'FCz', 'FC2', 'FC4', 'FC6', 'C5', 'C3', 'C1', 'Cz', 'C6', 'CP5', 'CP3', 'CP1', 'Fp1', 'Fpz', 'Fp2', 'AF7', 'AF3', 'AFz', 'AF4', 'AF8', 'F7', 'F5', 'F3', 'F1', 'Fz', 'F2', 'F4', 'F6', 'F8', 'FT7', 'FT8', 'T7', 'T8', 'T9', 'T10', 'TP7', 'TP8', 'P7', 'P5', 'P3', 'P1', 'Pz', 'P2', 'P4', 'P6', 'P8', 'PO7', 'PO3', 'POz', 'PO4', 'PO8', 'O1', 'Oz', 'O2', 'Iz']
Rejecting epoch based on EEG : ['Fp1', 'Fpz', 'Fp2', 'AF7', 'AF3', 'AF8']
Rejecting epoch based on EEG : ['Fp1', 'Fpz', 'Fp2', 'AF7', 'AF3']
Rejecting epoch based on EEG : ['Fp1', 'Fpz', 'Fp2', 'AF7', 'AF3']
Rejecting epoch based on EEG : ['Fp1']
Rejecting epoch based on EEG : ['Fp1', 'Fpz', 'Fp2', 'AF7', 'AF3', 'AF8']
Rejecting epoch based on EEG : ['Fp1']
Rejecting epoch based on EEG : ['Fp1']
10 bad epochs dropped
Setting up high-pass filter at 1 Hz
FIR filter parameters
---------------------
Designing a one-pass, zero-phase, non-causal highpass filter:
- Windowed time-domain design (firwin) method
- Hamming window with 0.0194 passband ripple and 53 dB stopband attenuation
- Lower passband edge: 1.00
- Lower transition bandwidth: 1.00 Hz (-6 dB cutoff frequency: 0.50 Hz)
- Filter length: 529 samples (3.306 s)
Fitting ICA to data using 64 channels (please be patient, this may take a while)
Selecting by number: 15 components
Fitting ICA took 0.5s.
Using EOG channel: Fp1
Using multitaper spectrum estimation with 7 DPSS windows
Applying ICA to Epochs instance
Transforming to ICA space (15 components)
Zeroing out 2 ICA components
Projecting back using 64 PCA components
Source space : <SourceSpaces: [<volume, shape=(np.int32(33), np.int32(39), np.int32(34)), n_used=14629>] MRI (surface RAS) coords, subject 'fsaverage', ~75.3 MiB>
MRI -> head transform : /home/circleci/project/mne-python/mne/data/fsaverage/fsaverage-trans.fif
Measurement data : instance of Info
Conductor model : instance of ConductorModel
Accurate field computations
Do computations in head coordinates
Free source orientations
Read 1 source spaces a total of 14629 active source locations
Coordinate transformation: MRI (surface RAS) -> head
0.999994 0.003552 0.000202 -1.76 mm
-0.003558 0.998389 0.056626 31.09 mm
-0.000001 -0.056626 0.998395 39.60 mm
0.000000 0.000000 0.000000 1.00
Read 64 EEG channels from info
Head coordinate coil definitions created.
Source spaces are now in head coordinates.
Employing the head->MRI coordinate transform with the BEM model.
BEM model instance of ConductorModel is now set up
Source spaces are in head coordinates.
Checking that the sources are inside the surface and at least 5.0 mm away (will take a few...)
Checking surface interior status for 14629 points...
Found 3644/14629 points inside an interior sphere of radius 47.7 mm
Found 0/14629 points outside an exterior sphere of radius 98.3 mm
Found 0/10985 points outside using surface Qhull
Found 0/10985 points outside using solid angles
Total 14629/14629 points inside the surface
Interior check completed in 10534.7 ms
Setting up for EEG...
Computing EEG at 14629 source locations (free orientations)...
Finished.
Computing rank from data with rank=None
Estimated rank (eeg): 63
EEG: rank 63 computed from 64 data channels with 1 projector
NOTE: tfr_morlet() is a legacy function. New code should use .compute_tfr(method="morlet").
/home/circleci/project/examples/volume_stc_group_study.py:124: RuntimeWarning: The measurement information indicates a low-pass frequency of 80.0 Hz. The decim=20 parameter will result in a sampling frequency of 8.0 Hz, which can cause aliasing artifacts.
epochs.decimate(20)
Created an SSP operator (subspace dimension = 1)
Setting small EEG eigenvalues to zero (without PCA)
Reducing data rank from 64 -> 63
Estimating covariance using EMPIRICAL
Done.
Number of samples used : 455
[done]
Computing inverse operator with 64 channels.
64 out of 64 channels remain after picking
Selected 64 channels
Creating the depth weighting matrix...
64 EEG channels
limit = 14630/14629 = 4.165017
scale = 49111.7 exp = 0.8
Whitening the forward solution.
Created an SSP operator (subspace dimension = 1)
Computing rank from covariance with rank=None
Using tolerance 1.4e-12 (2.2e-16 eps * 64 dim * 99 max singular value)
Estimated rank (eeg): 63
EEG: rank 63 computed from 64 data channels with 1 projector
Setting small EEG eigenvalues to zero (without PCA)
Creating the source covariance matrix
Adjusting source covariance matrix.
Computing SVD of whitened and weighted lead field matrix.
largest singular value = 7.91938
scaling factor to adjust the trace = 1.94998e+21 (nchan = 64 nzero = 1)
Computing inverse operator with 64 channels.
64 out of 64 channels remain after picking
Selected 64 channels
Creating the depth weighting matrix...
64 EEG channels
limit = 14630/14629 = 4.165017
scale = 49111.7 exp = 0.8
Whitening the forward solution.
Created an SSP operator (subspace dimension = 1)
Computing rank from covariance with rank=None
Using tolerance 7.6e-14 (2.2e-16 eps * 64 dim * 5.3 max singular value)
Estimated rank (eeg): 62
EEG: rank 62 computed from 64 data channels with 1 projector
Setting small EEG eigenvalues to zero (without PCA)
Creating the source covariance matrix
Adjusting source covariance matrix.
Computing SVD of whitened and weighted lead field matrix.
largest singular value = 4.8247
scaling factor to adjust the trace = 9.85846e+20 (nchan = 64 nzero = 2)
Computing inverse operator with 64 channels.
64 out of 64 channels remain after picking
Selected 64 channels
Creating the depth weighting matrix...
64 EEG channels
limit = 14630/14629 = 4.165017
scale = 49111.7 exp = 0.8
Whitening the forward solution.
Created an SSP operator (subspace dimension = 1)
Computing rank from covariance with rank=None
Using tolerance 6.4e-14 (2.2e-16 eps * 64 dim * 4.5 max singular value)
Estimated rank (eeg): 62
EEG: rank 62 computed from 64 data channels with 1 projector
Setting small EEG eigenvalues to zero (without PCA)
Creating the source covariance matrix
Adjusting source covariance matrix.
Computing SVD of whitened and weighted lead field matrix.
largest singular value = 4.78068
scaling factor to adjust the trace = 1.09743e+21 (nchan = 64 nzero = 2)
Computing inverse operator with 64 channels.
64 out of 64 channels remain after picking
Selected 64 channels
Creating the depth weighting matrix...
64 EEG channels
limit = 14630/14629 = 4.165017
scale = 49111.7 exp = 0.8
Whitening the forward solution.
Created an SSP operator (subspace dimension = 1)
Computing rank from covariance with rank=None
Using tolerance 2.9e-14 (2.2e-16 eps * 64 dim * 2.1 max singular value)
Estimated rank (eeg): 62
EEG: rank 62 computed from 64 data channels with 1 projector
Setting small EEG eigenvalues to zero (without PCA)
Creating the source covariance matrix
Adjusting source covariance matrix.
Computing SVD of whitened and weighted lead field matrix.
largest singular value = 4.97082
scaling factor to adjust the trace = 1.29189e+21 (nchan = 64 nzero = 2)
Computing inverse operator with 64 channels.
64 out of 64 channels remain after picking
Selected 64 channels
Creating the depth weighting matrix...
64 EEG channels
limit = 14630/14629 = 4.165017
scale = 49111.7 exp = 0.8
Whitening the forward solution.
Created an SSP operator (subspace dimension = 1)
Computing rank from covariance with rank=None
Using tolerance 1.6e-14 (2.2e-16 eps * 64 dim * 1.1 max singular value)
Estimated rank (eeg): 62
EEG: rank 62 computed from 64 data channels with 1 projector
Setting small EEG eigenvalues to zero (without PCA)
Creating the source covariance matrix
Adjusting source covariance matrix.
Computing SVD of whitened and weighted lead field matrix.
largest singular value = 5.37728
scaling factor to adjust the trace = 1.61299e+21 (nchan = 64 nzero = 2)
Computing inverse operator with 64 channels.
64 out of 64 channels remain after picking
Selected 64 channels
Creating the depth weighting matrix...
64 EEG channels
limit = 14630/14629 = 4.165017
scale = 49111.7 exp = 0.8
Whitening the forward solution.
Created an SSP operator (subspace dimension = 1)
Computing rank from covariance with rank=None
Using tolerance 1.4e-14 (2.2e-16 eps * 64 dim * 1 max singular value)
Estimated rank (eeg): 62
EEG: rank 62 computed from 64 data channels with 1 projector
Setting small EEG eigenvalues to zero (without PCA)
Creating the source covariance matrix
Adjusting source covariance matrix.
Computing SVD of whitened and weighted lead field matrix.
largest singular value = 5.49946
scaling factor to adjust the trace = 1.82681e+21 (nchan = 64 nzero = 2)
Computing inverse operator with 64 channels.
64 out of 64 channels remain after picking
Selected 64 channels
Creating the depth weighting matrix...
64 EEG channels
limit = 14630/14629 = 4.165017
scale = 49111.7 exp = 0.8
Whitening the forward solution.
Created an SSP operator (subspace dimension = 1)
Computing rank from covariance with rank=None
Using tolerance 1.3e-14 (2.2e-16 eps * 64 dim * 0.91 max singular value)
Estimated rank (eeg): 62
EEG: rank 62 computed from 64 data channels with 1 projector
Setting small EEG eigenvalues to zero (without PCA)
Creating the source covariance matrix
Adjusting source covariance matrix.
Computing SVD of whitened and weighted lead field matrix.
largest singular value = 5.7522
scaling factor to adjust the trace = 2.23206e+21 (nchan = 64 nzero = 2)
Computing inverse operator with 64 channels.
64 out of 64 channels remain after picking
Selected 64 channels
Creating the depth weighting matrix...
64 EEG channels
limit = 14630/14629 = 4.165017
scale = 49111.7 exp = 0.8
Whitening the forward solution.
Created an SSP operator (subspace dimension = 1)
Computing rank from covariance with rank=None
Using tolerance 1.2e-14 (2.2e-16 eps * 64 dim * 0.83 max singular value)
Estimated rank (eeg): 62
EEG: rank 62 computed from 64 data channels with 1 projector
Setting small EEG eigenvalues to zero (without PCA)
Creating the source covariance matrix
Adjusting source covariance matrix.
Computing SVD of whitened and weighted lead field matrix.
largest singular value = 6.11807
scaling factor to adjust the trace = 2.81508e+21 (nchan = 64 nzero = 2)
Computing inverse operator with 64 channels.
64 out of 64 channels remain after picking
Selected 64 channels
Creating the depth weighting matrix...
64 EEG channels
limit = 14630/14629 = 4.165017
scale = 49111.7 exp = 0.8
Whitening the forward solution.
Created an SSP operator (subspace dimension = 1)
Computing rank from covariance with rank=None
Using tolerance 8.2e-15 (2.2e-16 eps * 64 dim * 0.58 max singular value)
Estimated rank (eeg): 62
EEG: rank 62 computed from 64 data channels with 1 projector
Setting small EEG eigenvalues to zero (without PCA)
Creating the source covariance matrix
Adjusting source covariance matrix.
Computing SVD of whitened and weighted lead field matrix.
largest singular value = 6.12595
scaling factor to adjust the trace = 3.30957e+21 (nchan = 64 nzero = 2)
Computing inverse operator with 64 channels.
64 out of 64 channels remain after picking
Selected 64 channels
Creating the depth weighting matrix...
64 EEG channels
limit = 14630/14629 = 4.165017
scale = 49111.7 exp = 0.8
Whitening the forward solution.
Created an SSP operator (subspace dimension = 1)
Computing rank from covariance with rank=None
Using tolerance 4.8e-15 (2.2e-16 eps * 64 dim * 0.34 max singular value)
Estimated rank (eeg): 62
EEG: rank 62 computed from 64 data channels with 1 projector
Setting small EEG eigenvalues to zero (without PCA)
Creating the source covariance matrix
Adjusting source covariance matrix.
Computing SVD of whitened and weighted lead field matrix.
largest singular value = 5.99843
scaling factor to adjust the trace = 3.7062e+21 (nchan = 64 nzero = 2)
Computing source estimate for subject 2
Downloading EEGBCI data
Download complete in 18s (7.3 MB)
Used Annotations descriptions: [np.str_('T1'), np.str_('T2')]
Not setting metadata
45 matching events found
No baseline correction applied
0 projection items activated
Using data from preloaded Raw for 45 events and 961 original time points ...
3 bad epochs dropped
EEG channel type selected for re-referencing
Adding average EEG reference projection.
1 projection items deactivated
Average reference projection was added, but has not been applied yet. Use the apply_proj method to apply it.
Estimating rejection dictionary for eeg
Rejecting epoch based on EEG : ['Fp1', 'Fpz', 'Fp2', 'AF7', 'AF3', 'AFz', 'AF4', 'AF8']
Rejecting epoch based on EEG : ['Fp1', 'Fpz', 'Fp2', 'AF7', 'AF8']
Rejecting epoch based on EEG : ['FC5', 'FC3', 'FC1', 'FC4', 'FC6', 'C5', 'C3', 'C4', 'C6', 'CP5', 'CP6', 'Fp1', 'Fpz', 'AF7', 'F7', 'F5', 'F3', 'F6']
Rejecting epoch based on EEG : ['FC5', 'FC3', 'FC4', 'FC6', 'C5', 'C3', 'C4', 'C6', 'CP5', 'CP6', 'Fp1', 'Fpz', 'Fp2', 'AF7', 'AF8', 'F7', 'F5', 'F3', 'F4', 'F6', 'T7']
Rejecting epoch based on EEG : ['FC5', 'FC3', 'FC1', 'FC2', 'FC4', 'FC6', 'C5', 'C3', 'C4', 'C6', 'CP5', 'CP4', 'CP6', 'AF7', 'F7', 'F5', 'F6', 'F8', 'T7', 'T8', 'TP7']
Rejecting epoch based on EEG : ['FC5', 'FC3', 'C5', 'C3', 'C6', 'CP5', 'Fp1', 'AF7', 'F7', 'F5']
Rejecting epoch based on EEG : ['FC5', 'FC3', 'FC6', 'C5', 'C6', 'CP5', 'F7', 'F5', 'F3']
Rejecting epoch based on EEG : ['FC3', 'FC6', 'C5', 'C3', 'C6', 'CP5', 'CP3']
8 bad epochs dropped
Setting up high-pass filter at 1 Hz
FIR filter parameters
---------------------
Designing a one-pass, zero-phase, non-causal highpass filter:
- Windowed time-domain design (firwin) method
- Hamming window with 0.0194 passband ripple and 53 dB stopband attenuation
- Lower passband edge: 1.00
- Lower transition bandwidth: 1.00 Hz (-6 dB cutoff frequency: 0.50 Hz)
- Filter length: 529 samples (3.306 s)
Fitting ICA to data using 64 channels (please be patient, this may take a while)
Selecting by number: 15 components
Fitting ICA took 0.4s.
Using EOG channel: Fp1
Using multitaper spectrum estimation with 7 DPSS windows
Applying ICA to Epochs instance
Transforming to ICA space (15 components)
Zeroing out 5 ICA components
Projecting back using 64 PCA components
Source space : <SourceSpaces: [<volume, shape=(np.int32(33), np.int32(39), np.int32(34)), n_used=14629>] MRI (surface RAS) coords, subject 'fsaverage', ~75.3 MiB>
MRI -> head transform : /home/circleci/project/mne-python/mne/data/fsaverage/fsaverage-trans.fif
Measurement data : instance of Info
Conductor model : instance of ConductorModel
Accurate field computations
Do computations in head coordinates
Free source orientations
Read 1 source spaces a total of 14629 active source locations
Coordinate transformation: MRI (surface RAS) -> head
0.999994 0.003552 0.000202 -1.76 mm
-0.003558 0.998389 0.056626 31.09 mm
-0.000001 -0.056626 0.998395 39.60 mm
0.000000 0.000000 0.000000 1.00
Read 64 EEG channels from info
Head coordinate coil definitions created.
Source spaces are now in head coordinates.
Employing the head->MRI coordinate transform with the BEM model.
BEM model instance of ConductorModel is now set up
Source spaces are in head coordinates.
Checking that the sources are inside the surface and at least 5.0 mm away (will take a few...)
Checking surface interior status for 14629 points...
Found 3644/14629 points inside an interior sphere of radius 47.7 mm
Found 0/14629 points outside an exterior sphere of radius 98.3 mm
Found 0/10985 points outside using surface Qhull
Found 0/10985 points outside using solid angles
Total 14629/14629 points inside the surface
Interior check completed in 10774.2 ms
Setting up for EEG...
Computing EEG at 14629 source locations (free orientations)...
Finished.
Computing rank from data with rank=None
Estimated rank (eeg): 60
EEG: rank 60 computed from 64 data channels with 1 projector
NOTE: tfr_morlet() is a legacy function. New code should use .compute_tfr(method="morlet").
/home/circleci/project/examples/volume_stc_group_study.py:124: RuntimeWarning: The measurement information indicates a low-pass frequency of 80.0 Hz. The decim=20 parameter will result in a sampling frequency of 8.0 Hz, which can cause aliasing artifacts.
epochs.decimate(20)
Created an SSP operator (subspace dimension = 1)
Setting small EEG eigenvalues to zero (without PCA)
Reducing data rank from 64 -> 60
Estimating covariance using EMPIRICAL
Done.
Number of samples used : 442
[done]
Computing inverse operator with 64 channels.
64 out of 64 channels remain after picking
Selected 64 channels
Creating the depth weighting matrix...
64 EEG channels
limit = 14630/14629 = 4.165017
scale = 49111.7 exp = 0.8
Whitening the forward solution.
Created an SSP operator (subspace dimension = 1)
Computing rank from covariance with rank=None
Using tolerance 5.9e-13 (2.2e-16 eps * 64 dim * 42 max singular value)
Estimated rank (eeg): 60
EEG: rank 60 computed from 64 data channels with 1 projector
Setting small EEG eigenvalues to zero (without PCA)
Creating the source covariance matrix
Adjusting source covariance matrix.
Computing SVD of whitened and weighted lead field matrix.
largest singular value = 7.60856
scaling factor to adjust the trace = 6.02793e+20 (nchan = 64 nzero = 4)
Computing inverse operator with 64 channels.
64 out of 64 channels remain after picking
Selected 64 channels
Creating the depth weighting matrix...
64 EEG channels
limit = 14630/14629 = 4.165017
scale = 49111.7 exp = 0.8
Whitening the forward solution.
Created an SSP operator (subspace dimension = 1)
Computing rank from covariance with rank=None
Using tolerance 3.8e-14 (2.2e-16 eps * 64 dim * 2.7 max singular value)
Estimated rank (eeg): 59
EEG: rank 59 computed from 64 data channels with 1 projector
Setting small EEG eigenvalues to zero (without PCA)
Creating the source covariance matrix
Adjusting source covariance matrix.
Computing SVD of whitened and weighted lead field matrix.
largest singular value = 6.2783
scaling factor to adjust the trace = 1.6984e+21 (nchan = 64 nzero = 5)
Computing inverse operator with 64 channels.
64 out of 64 channels remain after picking
Selected 64 channels
Creating the depth weighting matrix...
64 EEG channels
limit = 14630/14629 = 4.165017
scale = 49111.7 exp = 0.8
Whitening the forward solution.
Created an SSP operator (subspace dimension = 1)
Computing rank from covariance with rank=None
Using tolerance 2.1e-14 (2.2e-16 eps * 64 dim * 1.5 max singular value)
Estimated rank (eeg): 59
EEG: rank 59 computed from 64 data channels with 1 projector
Setting small EEG eigenvalues to zero (without PCA)
Creating the source covariance matrix
Adjusting source covariance matrix.
Computing SVD of whitened and weighted lead field matrix.
largest singular value = 6.23316
scaling factor to adjust the trace = 1.89071e+21 (nchan = 64 nzero = 5)
Computing inverse operator with 64 channels.
64 out of 64 channels remain after picking
Selected 64 channels
Creating the depth weighting matrix...
64 EEG channels
limit = 14630/14629 = 4.165017
scale = 49111.7 exp = 0.8
Whitening the forward solution.
Created an SSP operator (subspace dimension = 1)
Computing rank from covariance with rank=None
Using tolerance 8.6e-15 (2.2e-16 eps * 64 dim * 0.6 max singular value)
Estimated rank (eeg): 59
EEG: rank 59 computed from 64 data channels with 1 projector
Setting small EEG eigenvalues to zero (without PCA)
Creating the source covariance matrix
Adjusting source covariance matrix.
Computing SVD of whitened and weighted lead field matrix.
largest singular value = 6.07464
scaling factor to adjust the trace = 2.01571e+21 (nchan = 64 nzero = 5)
Computing inverse operator with 64 channels.
64 out of 64 channels remain after picking
Selected 64 channels
Creating the depth weighting matrix...
64 EEG channels
limit = 14630/14629 = 4.165017
scale = 49111.7 exp = 0.8
Whitening the forward solution.
Created an SSP operator (subspace dimension = 1)
Computing rank from covariance with rank=None
Using tolerance 5.9e-15 (2.2e-16 eps * 64 dim * 0.42 max singular value)
Estimated rank (eeg): 59
EEG: rank 59 computed from 64 data channels with 1 projector
Setting small EEG eigenvalues to zero (without PCA)
Creating the source covariance matrix
Adjusting source covariance matrix.
Computing SVD of whitened and weighted lead field matrix.
largest singular value = 5.97538
scaling factor to adjust the trace = 1.97715e+21 (nchan = 64 nzero = 5)
Computing inverse operator with 64 channels.
64 out of 64 channels remain after picking
Selected 64 channels
Creating the depth weighting matrix...
64 EEG channels
limit = 14630/14629 = 4.165017
scale = 49111.7 exp = 0.8
Whitening the forward solution.
Created an SSP operator (subspace dimension = 1)
Computing rank from covariance with rank=None
Using tolerance 6.2e-15 (2.2e-16 eps * 64 dim * 0.44 max singular value)
Estimated rank (eeg): 59
EEG: rank 59 computed from 64 data channels with 1 projector
Setting small EEG eigenvalues to zero (without PCA)
Creating the source covariance matrix
Adjusting source covariance matrix.
Computing SVD of whitened and weighted lead field matrix.
largest singular value = 5.97412
scaling factor to adjust the trace = 1.80161e+21 (nchan = 64 nzero = 5)
Computing inverse operator with 64 channels.
64 out of 64 channels remain after picking
Selected 64 channels
Creating the depth weighting matrix...
64 EEG channels
limit = 14630/14629 = 4.165017
scale = 49111.7 exp = 0.8
Whitening the forward solution.
Created an SSP operator (subspace dimension = 1)
Computing rank from covariance with rank=None
Using tolerance 8e-15 (2.2e-16 eps * 64 dim * 0.57 max singular value)
Estimated rank (eeg): 59
EEG: rank 59 computed from 64 data channels with 1 projector
Setting small EEG eigenvalues to zero (without PCA)
Creating the source covariance matrix
Adjusting source covariance matrix.
Computing SVD of whitened and weighted lead field matrix.
largest singular value = 6.07409
scaling factor to adjust the trace = 1.81024e+21 (nchan = 64 nzero = 5)
Computing inverse operator with 64 channels.
64 out of 64 channels remain after picking
Selected 64 channels
Creating the depth weighting matrix...
64 EEG channels
limit = 14630/14629 = 4.165017
scale = 49111.7 exp = 0.8
Whitening the forward solution.
Created an SSP operator (subspace dimension = 1)
Computing rank from covariance with rank=None
Using tolerance 9.3e-15 (2.2e-16 eps * 64 dim * 0.65 max singular value)
Estimated rank (eeg): 59
EEG: rank 59 computed from 64 data channels with 1 projector
Setting small EEG eigenvalues to zero (without PCA)
Creating the source covariance matrix
Adjusting source covariance matrix.
Computing SVD of whitened and weighted lead field matrix.
largest singular value = 6.09589
scaling factor to adjust the trace = 2.0662e+21 (nchan = 64 nzero = 5)
Computing inverse operator with 64 channels.
64 out of 64 channels remain after picking
Selected 64 channels
Creating the depth weighting matrix...
64 EEG channels
limit = 14630/14629 = 4.165017
scale = 49111.7 exp = 0.8
Whitening the forward solution.
Created an SSP operator (subspace dimension = 1)
Computing rank from covariance with rank=None
Using tolerance 4.3e-15 (2.2e-16 eps * 64 dim * 0.3 max singular value)
Estimated rank (eeg): 59
EEG: rank 59 computed from 64 data channels with 1 projector
Setting small EEG eigenvalues to zero (without PCA)
Creating the source covariance matrix
Adjusting source covariance matrix.
Computing SVD of whitened and weighted lead field matrix.
largest singular value = 5.71597
scaling factor to adjust the trace = 2.35867e+21 (nchan = 64 nzero = 5)
Computing inverse operator with 64 channels.
64 out of 64 channels remain after picking
Selected 64 channels
Creating the depth weighting matrix...
64 EEG channels
limit = 14630/14629 = 4.165017
scale = 49111.7 exp = 0.8
Whitening the forward solution.
Created an SSP operator (subspace dimension = 1)
Computing rank from covariance with rank=None
Using tolerance 2.8e-15 (2.2e-16 eps * 64 dim * 0.2 max singular value)
Estimated rank (eeg): 59
EEG: rank 59 computed from 64 data channels with 1 projector
Setting small EEG eigenvalues to zero (without PCA)
Creating the source covariance matrix
Adjusting source covariance matrix.
Computing SVD of whitened and weighted lead field matrix.
largest singular value = 5.43242
scaling factor to adjust the trace = 2.59812e+21 (nchan = 64 nzero = 5)
Computing source estimate for subject 3
Downloading EEGBCI data
Download complete in 19s (7.4 MB)
Used Annotations descriptions: [np.str_('T1'), np.str_('T2')]
Not setting metadata
45 matching events found
No baseline correction applied
0 projection items activated
Using data from preloaded Raw for 45 events and 961 original time points ...
0 bad epochs dropped
EEG channel type selected for re-referencing
Adding average EEG reference projection.
1 projection items deactivated
Average reference projection was added, but has not been applied yet. Use the apply_proj method to apply it.
Estimating rejection dictionary for eeg
0 bad epochs dropped
Setting up high-pass filter at 1 Hz
FIR filter parameters
---------------------
Designing a one-pass, zero-phase, non-causal highpass filter:
- Windowed time-domain design (firwin) method
- Hamming window with 0.0194 passband ripple and 53 dB stopband attenuation
- Lower passband edge: 1.00
- Lower transition bandwidth: 1.00 Hz (-6 dB cutoff frequency: 0.50 Hz)
- Filter length: 529 samples (3.306 s)
Fitting ICA to data using 64 channels (please be patient, this may take a while)
Selecting by number: 15 components
Fitting ICA took 0.8s.
Using EOG channel: Fp1
Using multitaper spectrum estimation with 7 DPSS windows
Applying ICA to Epochs instance
Transforming to ICA space (15 components)
Zeroing out 7 ICA components
Projecting back using 64 PCA components
Source space : <SourceSpaces: [<volume, shape=(np.int32(33), np.int32(39), np.int32(34)), n_used=14629>] MRI (surface RAS) coords, subject 'fsaverage', ~75.3 MiB>
MRI -> head transform : /home/circleci/project/mne-python/mne/data/fsaverage/fsaverage-trans.fif
Measurement data : instance of Info
Conductor model : instance of ConductorModel
Accurate field computations
Do computations in head coordinates
Free source orientations
Read 1 source spaces a total of 14629 active source locations
Coordinate transformation: MRI (surface RAS) -> head
0.999994 0.003552 0.000202 -1.76 mm
-0.003558 0.998389 0.056626 31.09 mm
-0.000001 -0.056626 0.998395 39.60 mm
0.000000 0.000000 0.000000 1.00
Read 64 EEG channels from info
Head coordinate coil definitions created.
Source spaces are now in head coordinates.
Employing the head->MRI coordinate transform with the BEM model.
BEM model instance of ConductorModel is now set up
Source spaces are in head coordinates.
Checking that the sources are inside the surface and at least 5.0 mm away (will take a few...)
Checking surface interior status for 14629 points...
Found 3644/14629 points inside an interior sphere of radius 47.7 mm
Found 0/14629 points outside an exterior sphere of radius 98.3 mm
Found 0/10985 points outside using surface Qhull
Found 0/10985 points outside using solid angles
Total 14629/14629 points inside the surface
Interior check completed in 10078.5 ms
Setting up for EEG...
Computing EEG at 14629 source locations (free orientations)...
Finished.
Computing rank from data with rank=None
Estimated rank (eeg): 58
EEG: rank 58 computed from 64 data channels with 1 projector
NOTE: tfr_morlet() is a legacy function. New code should use .compute_tfr(method="morlet").
/home/circleci/project/examples/volume_stc_group_study.py:124: RuntimeWarning: The measurement information indicates a low-pass frequency of 80.0 Hz. The decim=20 parameter will result in a sampling frequency of 8.0 Hz, which can cause aliasing artifacts.
epochs.decimate(20)
Created an SSP operator (subspace dimension = 1)
Setting small EEG eigenvalues to zero (without PCA)
Reducing data rank from 64 -> 58
Estimating covariance using EMPIRICAL
Done.
Number of samples used : 585
[done]
Computing inverse operator with 64 channels.
64 out of 64 channels remain after picking
Selected 64 channels
Creating the depth weighting matrix...
64 EEG channels
limit = 14630/14629 = 4.165017
scale = 49111.7 exp = 0.8
Whitening the forward solution.
Created an SSP operator (subspace dimension = 1)
Computing rank from covariance with rank=None
Using tolerance 8.9e-13 (2.2e-16 eps * 64 dim * 63 max singular value)
Estimated rank (eeg): 58
EEG: rank 58 computed from 64 data channels with 1 projector
Setting small EEG eigenvalues to zero (without PCA)
Creating the source covariance matrix
Adjusting source covariance matrix.
Computing SVD of whitened and weighted lead field matrix.
largest singular value = 7.03207
scaling factor to adjust the trace = 9.42072e+19 (nchan = 64 nzero = 6)
Computing inverse operator with 64 channels.
64 out of 64 channels remain after picking
Selected 64 channels
Creating the depth weighting matrix...
64 EEG channels
limit = 14630/14629 = 4.165017
scale = 49111.7 exp = 0.8
Whitening the forward solution.
Created an SSP operator (subspace dimension = 1)
Computing rank from covariance with rank=None
Using tolerance 2.6e-14 (2.2e-16 eps * 64 dim * 1.9 max singular value)
Estimated rank (eeg): 57
EEG: rank 57 computed from 64 data channels with 1 projector
Setting small EEG eigenvalues to zero (without PCA)
Creating the source covariance matrix
Adjusting source covariance matrix.
Computing SVD of whitened and weighted lead field matrix.
largest singular value = 5.39417
scaling factor to adjust the trace = 1.78314e+21 (nchan = 64 nzero = 7)
Computing inverse operator with 64 channels.
64 out of 64 channels remain after picking
Selected 64 channels
Creating the depth weighting matrix...
64 EEG channels
limit = 14630/14629 = 4.165017
scale = 49111.7 exp = 0.8
Whitening the forward solution.
Created an SSP operator (subspace dimension = 1)
Computing rank from covariance with rank=None
Using tolerance 1.7e-14 (2.2e-16 eps * 64 dim * 1.2 max singular value)
Estimated rank (eeg): 57
EEG: rank 57 computed from 64 data channels with 1 projector
Setting small EEG eigenvalues to zero (without PCA)
Creating the source covariance matrix
Adjusting source covariance matrix.
Computing SVD of whitened and weighted lead field matrix.
largest singular value = 5.27789
scaling factor to adjust the trace = 1.95119e+21 (nchan = 64 nzero = 7)
Computing inverse operator with 64 channels.
64 out of 64 channels remain after picking
Selected 64 channels
Creating the depth weighting matrix...
64 EEG channels
limit = 14630/14629 = 4.165017
scale = 49111.7 exp = 0.8
Whitening the forward solution.
Created an SSP operator (subspace dimension = 1)
Computing rank from covariance with rank=None
Using tolerance 1.2e-14 (2.2e-16 eps * 64 dim * 0.86 max singular value)
Estimated rank (eeg): 57
EEG: rank 57 computed from 64 data channels with 1 projector
Setting small EEG eigenvalues to zero (without PCA)
Creating the source covariance matrix
Adjusting source covariance matrix.
Computing SVD of whitened and weighted lead field matrix.
largest singular value = 5.24071
scaling factor to adjust the trace = 2.09812e+21 (nchan = 64 nzero = 7)
Computing inverse operator with 64 channels.
64 out of 64 channels remain after picking
Selected 64 channels
Creating the depth weighting matrix...
64 EEG channels
limit = 14630/14629 = 4.165017
scale = 49111.7 exp = 0.8
Whitening the forward solution.
Created an SSP operator (subspace dimension = 1)
Computing rank from covariance with rank=None
Using tolerance 1e-14 (2.2e-16 eps * 64 dim * 0.71 max singular value)
Estimated rank (eeg): 57
EEG: rank 57 computed from 64 data channels with 1 projector
Setting small EEG eigenvalues to zero (without PCA)
Creating the source covariance matrix
Adjusting source covariance matrix.
Computing SVD of whitened and weighted lead field matrix.
largest singular value = 5.26196
scaling factor to adjust the trace = 2.2252e+21 (nchan = 64 nzero = 7)
Computing inverse operator with 64 channels.
64 out of 64 channels remain after picking
Selected 64 channels
Creating the depth weighting matrix...
64 EEG channels
limit = 14630/14629 = 4.165017
scale = 49111.7 exp = 0.8
Whitening the forward solution.
Created an SSP operator (subspace dimension = 1)
Computing rank from covariance with rank=None
Using tolerance 9.5e-15 (2.2e-16 eps * 64 dim * 0.67 max singular value)
Estimated rank (eeg): 57
EEG: rank 57 computed from 64 data channels with 1 projector
Setting small EEG eigenvalues to zero (without PCA)
Creating the source covariance matrix
Adjusting source covariance matrix.
Computing SVD of whitened and weighted lead field matrix.
largest singular value = 5.2304
scaling factor to adjust the trace = 2.32255e+21 (nchan = 64 nzero = 7)
Computing inverse operator with 64 channels.
64 out of 64 channels remain after picking
Selected 64 channels
Creating the depth weighting matrix...
64 EEG channels
limit = 14630/14629 = 4.165017
scale = 49111.7 exp = 0.8
Whitening the forward solution.
Created an SSP operator (subspace dimension = 1)
Computing rank from covariance with rank=None
Using tolerance 7.3e-15 (2.2e-16 eps * 64 dim * 0.51 max singular value)
Estimated rank (eeg): 57
EEG: rank 57 computed from 64 data channels with 1 projector
Setting small EEG eigenvalues to zero (without PCA)
Creating the source covariance matrix
Adjusting source covariance matrix.
Computing SVD of whitened and weighted lead field matrix.
largest singular value = 5.03465
scaling factor to adjust the trace = 2.36961e+21 (nchan = 64 nzero = 7)
Computing inverse operator with 64 channels.
64 out of 64 channels remain after picking
Selected 64 channels
Creating the depth weighting matrix...
64 EEG channels
limit = 14630/14629 = 4.165017
scale = 49111.7 exp = 0.8
Whitening the forward solution.
Created an SSP operator (subspace dimension = 1)
Computing rank from covariance with rank=None
Using tolerance 5.3e-15 (2.2e-16 eps * 64 dim * 0.37 max singular value)
Estimated rank (eeg): 57
EEG: rank 57 computed from 64 data channels with 1 projector
Setting small EEG eigenvalues to zero (without PCA)
Creating the source covariance matrix
Adjusting source covariance matrix.
Computing SVD of whitened and weighted lead field matrix.
largest singular value = 4.83149
scaling factor to adjust the trace = 2.45637e+21 (nchan = 64 nzero = 7)
Computing inverse operator with 64 channels.
64 out of 64 channels remain after picking
Selected 64 channels
Creating the depth weighting matrix...
64 EEG channels
limit = 14630/14629 = 4.165017
scale = 49111.7 exp = 0.8
Whitening the forward solution.
Created an SSP operator (subspace dimension = 1)
Computing rank from covariance with rank=None
Using tolerance 3.4e-15 (2.2e-16 eps * 64 dim * 0.24 max singular value)
Estimated rank (eeg): 57
EEG: rank 57 computed from 64 data channels with 1 projector
Setting small EEG eigenvalues to zero (without PCA)
Creating the source covariance matrix
Adjusting source covariance matrix.
Computing SVD of whitened and weighted lead field matrix.
largest singular value = 4.48074
scaling factor to adjust the trace = 2.73257e+21 (nchan = 64 nzero = 7)
Computing inverse operator with 64 channels.
64 out of 64 channels remain after picking
Selected 64 channels
Creating the depth weighting matrix...
64 EEG channels
limit = 14630/14629 = 4.165017
scale = 49111.7 exp = 0.8
Whitening the forward solution.
Created an SSP operator (subspace dimension = 1)
Computing rank from covariance with rank=None
Using tolerance 2.1e-15 (2.2e-16 eps * 64 dim * 0.15 max singular value)
Estimated rank (eeg): 57
EEG: rank 57 computed from 64 data channels with 1 projector
Setting small EEG eigenvalues to zero (without PCA)
Creating the source covariance matrix
Adjusting source covariance matrix.
Computing SVD of whitened and weighted lead field matrix.
largest singular value = 4.45119
scaling factor to adjust the trace = 3.07374e+21 (nchan = 64 nzero = 7)
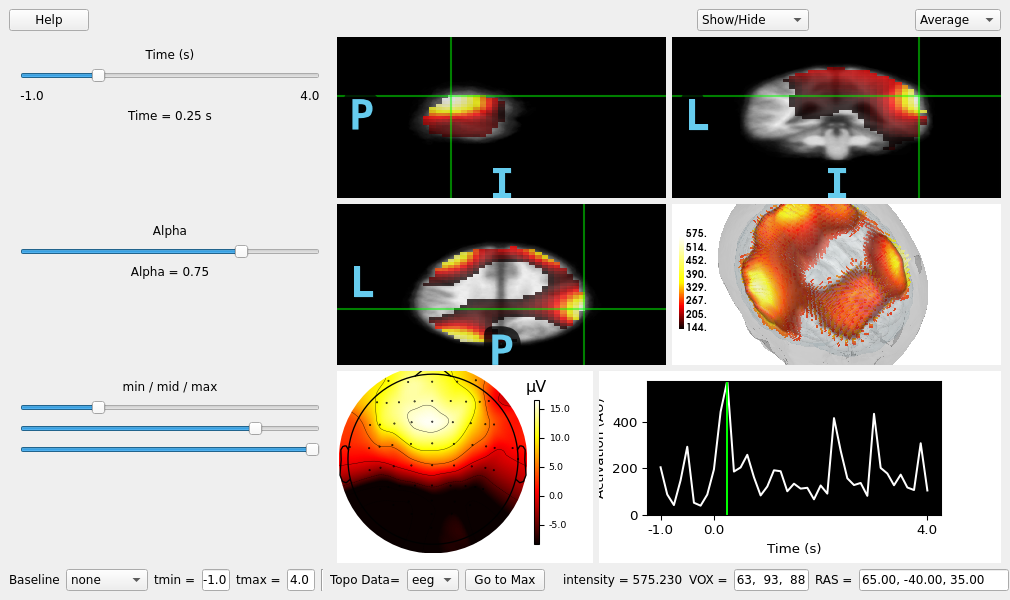
Use the viewer to explore the time-course source estimates.
viewer = mne_gui.view_vol_stc(
stcs_epochs,
group=True,
freq_first=False,
subject=template,
subjects_dir=subjects_dir,
src=src,
inst=insts_epochs,
tmin=tmin,
tmax=tmax,
)
viewer.go_to_extreme() # show the maximum intensity source vertex
viewer.set_cmap(vmin=0.25, vmid=0.8)
viewer.set_3d_view(azimuth=40, elevation=35, distance=300)
del stcs_epochs, insts_epochs
Preparing the inverse operator for use...
Scaled noise and source covariance from nave = 1 to nave = 1
Created the regularized inverter
Created an SSP operator (subspace dimension = 1)
Created the whitener using a noise covariance matrix with rank 63 (1 small eigenvalues omitted)
Picked 64 channels from the data
Computing inverse...
Eigenleads need to be weighted ...
Processing epoch : 1 / 35
Processing epoch : 2 / 35
Processing epoch : 3 / 35
Processing epoch : 4 / 35
Processing epoch : 5 / 35
Processing epoch : 6 / 35
Processing epoch : 7 / 35
Processing epoch : 8 / 35
Processing epoch : 9 / 35
Processing epoch : 10 / 35
Processing epoch : 11 / 35
Processing epoch : 12 / 35
Processing epoch : 13 / 35
Processing epoch : 14 / 35
Processing epoch : 15 / 35
Processing epoch : 16 / 35
Processing epoch : 17 / 35
Processing epoch : 18 / 35
Processing epoch : 19 / 35
Processing epoch : 20 / 35
Processing epoch : 21 / 35
Processing epoch : 22 / 35
Processing epoch : 23 / 35
Processing epoch : 24 / 35
Processing epoch : 25 / 35
Processing epoch : 26 / 35
Processing epoch : 27 / 35
Processing epoch : 28 / 35
Processing epoch : 29 / 35
Processing epoch : 30 / 35
Processing epoch : 31 / 35
Processing epoch : 32 / 35
Processing epoch : 33 / 35
Processing epoch : 34 / 35
Processing epoch : 35 / 35
[done]
Preparing the inverse operator for use...
Scaled noise and source covariance from nave = 1 to nave = 1
Created the regularized inverter
Created an SSP operator (subspace dimension = 1)
Created the whitener using a noise covariance matrix with rank 60 (4 small eigenvalues omitted)
Picked 64 channels from the data
Computing inverse...
Eigenleads need to be weighted ...
Processing epoch : 1 / 34
Processing epoch : 2 / 34
Processing epoch : 3 / 34
Processing epoch : 4 / 34
Processing epoch : 5 / 34
Processing epoch : 6 / 34
Processing epoch : 7 / 34
Processing epoch : 8 / 34
Processing epoch : 9 / 34
Processing epoch : 10 / 34
Processing epoch : 11 / 34
Processing epoch : 12 / 34
Processing epoch : 13 / 34
Processing epoch : 14 / 34
Processing epoch : 15 / 34
Processing epoch : 16 / 34
Processing epoch : 17 / 34
Processing epoch : 18 / 34
Processing epoch : 19 / 34
Processing epoch : 20 / 34
Processing epoch : 21 / 34
Processing epoch : 22 / 34
Processing epoch : 23 / 34
Processing epoch : 24 / 34
Processing epoch : 25 / 34
Processing epoch : 26 / 34
Processing epoch : 27 / 34
Processing epoch : 28 / 34
Processing epoch : 29 / 34
Processing epoch : 30 / 34
Processing epoch : 31 / 34
Processing epoch : 32 / 34
Processing epoch : 33 / 34
Processing epoch : 34 / 34
[done]
Preparing the inverse operator for use...
Scaled noise and source covariance from nave = 1 to nave = 1
Created the regularized inverter
Created an SSP operator (subspace dimension = 1)
Created the whitener using a noise covariance matrix with rank 58 (6 small eigenvalues omitted)
Picked 64 channels from the data
Computing inverse...
Eigenleads need to be weighted ...
Processing epoch : 1 / 45
Processing epoch : 2 / 45
Processing epoch : 3 / 45
Processing epoch : 4 / 45
Processing epoch : 5 / 45
Processing epoch : 6 / 45
Processing epoch : 7 / 45
Processing epoch : 8 / 45
Processing epoch : 9 / 45
Processing epoch : 10 / 45
Processing epoch : 11 / 45
Processing epoch : 12 / 45
Processing epoch : 13 / 45
Processing epoch : 14 / 45
Processing epoch : 15 / 45
Processing epoch : 16 / 45
Processing epoch : 17 / 45
Processing epoch : 18 / 45
Processing epoch : 19 / 45
Processing epoch : 20 / 45
Processing epoch : 21 / 45
Processing epoch : 22 / 45
Processing epoch : 23 / 45
Processing epoch : 24 / 45
Processing epoch : 25 / 45
Processing epoch : 26 / 45
Processing epoch : 27 / 45
Processing epoch : 28 / 45
Processing epoch : 29 / 45
Processing epoch : 30 / 45
Processing epoch : 31 / 45
Processing epoch : 32 / 45
Processing epoch : 33 / 45
Processing epoch : 34 / 45
Processing epoch : 35 / 45
Processing epoch : 36 / 45
Processing epoch : 37 / 45
Processing epoch : 38 / 45
Processing epoch : 39 / 45
Processing epoch : 40 / 45
Processing epoch : 41 / 45
Processing epoch : 42 / 45
Processing epoch : 43 / 45
Processing epoch : 44 / 45
Processing epoch : 45 / 45
[done]
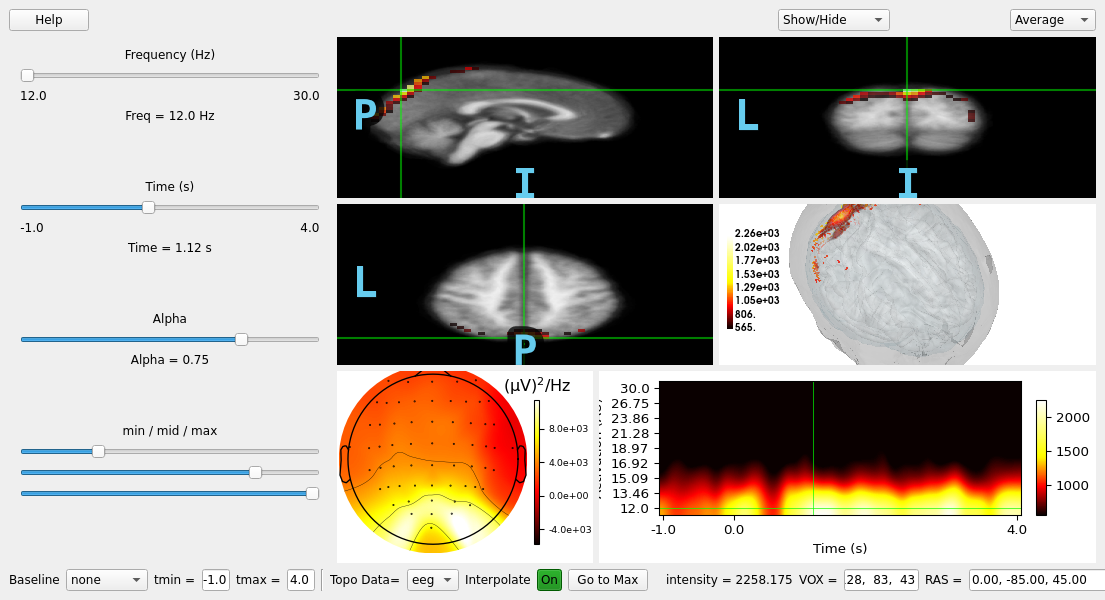
Use the viewer to explore the time-frequency source estimates, we’ll use the power in this case but you can also view inter-trial coherence (itc).
viewer = mne_gui.view_vol_stc(
stcs_tfr,
group="power", # can also be "itc"
subject=template,
subjects_dir=subjects_dir,
src=src,
inst=insts_tfr,
tmin=tmin,
tmax=tmax,
)
viewer.go_to_extreme() # show the maximum intensity source vertex
viewer.set_cmap(vmin=0.25, vmid=0.8)
viewer.set_3d_view(azimuth=40, elevation=35, distance=300)
del stcs_tfr, insts_tfr
Not setting metadata
35 matching events found
No baseline correction applied
Created an SSP operator (subspace dimension = 1)
1 projection items activated
Preparing the inverse operator for use...
Scaled noise and source covariance from nave = 1 to nave = 1
Created the regularized inverter
Created an SSP operator (subspace dimension = 1)
Created the whitener using a noise covariance matrix with rank 62 (2 small eigenvalues omitted)
Picked 64 channels from the data
Computing inverse...
Eigenleads need to be weighted ...
Processing epoch : 1 / 35
Processing epoch : 2 / 35
Processing epoch : 3 / 35
Processing epoch : 4 / 35
Processing epoch : 5 / 35
Processing epoch : 6 / 35
Processing epoch : 7 / 35
Processing epoch : 8 / 35
Processing epoch : 9 / 35
Processing epoch : 10 / 35
Processing epoch : 11 / 35
Processing epoch : 12 / 35
Processing epoch : 13 / 35
Processing epoch : 14 / 35
Processing epoch : 15 / 35
Processing epoch : 16 / 35
Processing epoch : 17 / 35
Processing epoch : 18 / 35
Processing epoch : 19 / 35
Processing epoch : 20 / 35
Processing epoch : 21 / 35
Processing epoch : 22 / 35
Processing epoch : 23 / 35
Processing epoch : 24 / 35
Processing epoch : 25 / 35
Processing epoch : 26 / 35
Processing epoch : 27 / 35
Processing epoch : 28 / 35
Processing epoch : 29 / 35
Processing epoch : 30 / 35
Processing epoch : 31 / 35
Processing epoch : 32 / 35
Processing epoch : 33 / 35
Processing epoch : 34 / 35
Processing epoch : 35 / 35
[done]
Not setting metadata
35 matching events found
No baseline correction applied
Created an SSP operator (subspace dimension = 1)
1 projection items activated
Preparing the inverse operator for use...
Scaled noise and source covariance from nave = 1 to nave = 1
Created the regularized inverter
Created an SSP operator (subspace dimension = 1)
Created the whitener using a noise covariance matrix with rank 62 (2 small eigenvalues omitted)
Picked 64 channels from the data
Computing inverse...
Eigenleads need to be weighted ...
Processing epoch : 1 / 35
Processing epoch : 2 / 35
Processing epoch : 3 / 35
Processing epoch : 4 / 35
Processing epoch : 5 / 35
Processing epoch : 6 / 35
Processing epoch : 7 / 35
Processing epoch : 8 / 35
Processing epoch : 9 / 35
Processing epoch : 10 / 35
Processing epoch : 11 / 35
Processing epoch : 12 / 35
Processing epoch : 13 / 35
Processing epoch : 14 / 35
Processing epoch : 15 / 35
Processing epoch : 16 / 35
Processing epoch : 17 / 35
Processing epoch : 18 / 35
Processing epoch : 19 / 35
Processing epoch : 20 / 35
Processing epoch : 21 / 35
Processing epoch : 22 / 35
Processing epoch : 23 / 35
Processing epoch : 24 / 35
Processing epoch : 25 / 35
Processing epoch : 26 / 35
Processing epoch : 27 / 35
Processing epoch : 28 / 35
Processing epoch : 29 / 35
Processing epoch : 30 / 35
Processing epoch : 31 / 35
Processing epoch : 32 / 35
Processing epoch : 33 / 35
Processing epoch : 34 / 35
Processing epoch : 35 / 35
[done]
Not setting metadata
35 matching events found
No baseline correction applied
Created an SSP operator (subspace dimension = 1)
1 projection items activated
Preparing the inverse operator for use...
Scaled noise and source covariance from nave = 1 to nave = 1
Created the regularized inverter
Created an SSP operator (subspace dimension = 1)
Created the whitener using a noise covariance matrix with rank 62 (2 small eigenvalues omitted)
Picked 64 channels from the data
Computing inverse...
Eigenleads need to be weighted ...
Processing epoch : 1 / 35
Processing epoch : 2 / 35
Processing epoch : 3 / 35
Processing epoch : 4 / 35
Processing epoch : 5 / 35
Processing epoch : 6 / 35
Processing epoch : 7 / 35
Processing epoch : 8 / 35
Processing epoch : 9 / 35
Processing epoch : 10 / 35
Processing epoch : 11 / 35
Processing epoch : 12 / 35
Processing epoch : 13 / 35
Processing epoch : 14 / 35
Processing epoch : 15 / 35
Processing epoch : 16 / 35
Processing epoch : 17 / 35
Processing epoch : 18 / 35
Processing epoch : 19 / 35
Processing epoch : 20 / 35
Processing epoch : 21 / 35
Processing epoch : 22 / 35
Processing epoch : 23 / 35
Processing epoch : 24 / 35
Processing epoch : 25 / 35
Processing epoch : 26 / 35
Processing epoch : 27 / 35
Processing epoch : 28 / 35
Processing epoch : 29 / 35
Processing epoch : 30 / 35
Processing epoch : 31 / 35
Processing epoch : 32 / 35
Processing epoch : 33 / 35
Processing epoch : 34 / 35
Processing epoch : 35 / 35
[done]
Not setting metadata
35 matching events found
No baseline correction applied
Created an SSP operator (subspace dimension = 1)
1 projection items activated
Preparing the inverse operator for use...
Scaled noise and source covariance from nave = 1 to nave = 1
Created the regularized inverter
Created an SSP operator (subspace dimension = 1)
Created the whitener using a noise covariance matrix with rank 62 (2 small eigenvalues omitted)
Picked 64 channels from the data
Computing inverse...
Eigenleads need to be weighted ...
Processing epoch : 1 / 35
Processing epoch : 2 / 35
Processing epoch : 3 / 35
Processing epoch : 4 / 35
Processing epoch : 5 / 35
Processing epoch : 6 / 35
Processing epoch : 7 / 35
Processing epoch : 8 / 35
Processing epoch : 9 / 35
Processing epoch : 10 / 35
Processing epoch : 11 / 35
Processing epoch : 12 / 35
Processing epoch : 13 / 35
Processing epoch : 14 / 35
Processing epoch : 15 / 35
Processing epoch : 16 / 35
Processing epoch : 17 / 35
Processing epoch : 18 / 35
Processing epoch : 19 / 35
Processing epoch : 20 / 35
Processing epoch : 21 / 35
Processing epoch : 22 / 35
Processing epoch : 23 / 35
Processing epoch : 24 / 35
Processing epoch : 25 / 35
Processing epoch : 26 / 35
Processing epoch : 27 / 35
Processing epoch : 28 / 35
Processing epoch : 29 / 35
Processing epoch : 30 / 35
Processing epoch : 31 / 35
Processing epoch : 32 / 35
Processing epoch : 33 / 35
Processing epoch : 34 / 35
Processing epoch : 35 / 35
[done]
Not setting metadata
35 matching events found
No baseline correction applied
Created an SSP operator (subspace dimension = 1)
1 projection items activated
Preparing the inverse operator for use...
Scaled noise and source covariance from nave = 1 to nave = 1
Created the regularized inverter
Created an SSP operator (subspace dimension = 1)
Created the whitener using a noise covariance matrix with rank 62 (2 small eigenvalues omitted)
Picked 64 channels from the data
Computing inverse...
Eigenleads need to be weighted ...
Processing epoch : 1 / 35
Processing epoch : 2 / 35
Processing epoch : 3 / 35
Processing epoch : 4 / 35
Processing epoch : 5 / 35
Processing epoch : 6 / 35
Processing epoch : 7 / 35
Processing epoch : 8 / 35
Processing epoch : 9 / 35
Processing epoch : 10 / 35
Processing epoch : 11 / 35
Processing epoch : 12 / 35
Processing epoch : 13 / 35
Processing epoch : 14 / 35
Processing epoch : 15 / 35
Processing epoch : 16 / 35
Processing epoch : 17 / 35
Processing epoch : 18 / 35
Processing epoch : 19 / 35
Processing epoch : 20 / 35
Processing epoch : 21 / 35
Processing epoch : 22 / 35
Processing epoch : 23 / 35
Processing epoch : 24 / 35
Processing epoch : 25 / 35
Processing epoch : 26 / 35
Processing epoch : 27 / 35
Processing epoch : 28 / 35
Processing epoch : 29 / 35
Processing epoch : 30 / 35
Processing epoch : 31 / 35
Processing epoch : 32 / 35
Processing epoch : 33 / 35
Processing epoch : 34 / 35
Processing epoch : 35 / 35
[done]
Not setting metadata
35 matching events found
No baseline correction applied
Created an SSP operator (subspace dimension = 1)
1 projection items activated
Preparing the inverse operator for use...
Scaled noise and source covariance from nave = 1 to nave = 1
Created the regularized inverter
Created an SSP operator (subspace dimension = 1)
Created the whitener using a noise covariance matrix with rank 62 (2 small eigenvalues omitted)
Picked 64 channels from the data
Computing inverse...
Eigenleads need to be weighted ...
Processing epoch : 1 / 35
Processing epoch : 2 / 35
Processing epoch : 3 / 35
Processing epoch : 4 / 35
Processing epoch : 5 / 35
Processing epoch : 6 / 35
Processing epoch : 7 / 35
Processing epoch : 8 / 35
Processing epoch : 9 / 35
Processing epoch : 10 / 35
Processing epoch : 11 / 35
Processing epoch : 12 / 35
Processing epoch : 13 / 35
Processing epoch : 14 / 35
Processing epoch : 15 / 35
Processing epoch : 16 / 35
Processing epoch : 17 / 35
Processing epoch : 18 / 35
Processing epoch : 19 / 35
Processing epoch : 20 / 35
Processing epoch : 21 / 35
Processing epoch : 22 / 35
Processing epoch : 23 / 35
Processing epoch : 24 / 35
Processing epoch : 25 / 35
Processing epoch : 26 / 35
Processing epoch : 27 / 35
Processing epoch : 28 / 35
Processing epoch : 29 / 35
Processing epoch : 30 / 35
Processing epoch : 31 / 35
Processing epoch : 32 / 35
Processing epoch : 33 / 35
Processing epoch : 34 / 35
Processing epoch : 35 / 35
[done]
Not setting metadata
35 matching events found
No baseline correction applied
Created an SSP operator (subspace dimension = 1)
1 projection items activated
Preparing the inverse operator for use...
Scaled noise and source covariance from nave = 1 to nave = 1
Created the regularized inverter
Created an SSP operator (subspace dimension = 1)
Created the whitener using a noise covariance matrix with rank 62 (2 small eigenvalues omitted)
Picked 64 channels from the data
Computing inverse...
Eigenleads need to be weighted ...
Processing epoch : 1 / 35
Processing epoch : 2 / 35
Processing epoch : 3 / 35
Processing epoch : 4 / 35
Processing epoch : 5 / 35
Processing epoch : 6 / 35
Processing epoch : 7 / 35
Processing epoch : 8 / 35
Processing epoch : 9 / 35
Processing epoch : 10 / 35
Processing epoch : 11 / 35
Processing epoch : 12 / 35
Processing epoch : 13 / 35
Processing epoch : 14 / 35
Processing epoch : 15 / 35
Processing epoch : 16 / 35
Processing epoch : 17 / 35
Processing epoch : 18 / 35
Processing epoch : 19 / 35
Processing epoch : 20 / 35
Processing epoch : 21 / 35
Processing epoch : 22 / 35
Processing epoch : 23 / 35
Processing epoch : 24 / 35
Processing epoch : 25 / 35
Processing epoch : 26 / 35
Processing epoch : 27 / 35
Processing epoch : 28 / 35
Processing epoch : 29 / 35
Processing epoch : 30 / 35
Processing epoch : 31 / 35
Processing epoch : 32 / 35
Processing epoch : 33 / 35
Processing epoch : 34 / 35
Processing epoch : 35 / 35
[done]
Not setting metadata
35 matching events found
No baseline correction applied
Created an SSP operator (subspace dimension = 1)
1 projection items activated
Preparing the inverse operator for use...
Scaled noise and source covariance from nave = 1 to nave = 1
Created the regularized inverter
Created an SSP operator (subspace dimension = 1)
Created the whitener using a noise covariance matrix with rank 62 (2 small eigenvalues omitted)
Picked 64 channels from the data
Computing inverse...
Eigenleads need to be weighted ...
Processing epoch : 1 / 35
Processing epoch : 2 / 35
Processing epoch : 3 / 35
Processing epoch : 4 / 35
Processing epoch : 5 / 35
Processing epoch : 6 / 35
Processing epoch : 7 / 35
Processing epoch : 8 / 35
Processing epoch : 9 / 35
Processing epoch : 10 / 35
Processing epoch : 11 / 35
Processing epoch : 12 / 35
Processing epoch : 13 / 35
Processing epoch : 14 / 35
Processing epoch : 15 / 35
Processing epoch : 16 / 35
Processing epoch : 17 / 35
Processing epoch : 18 / 35
Processing epoch : 19 / 35
Processing epoch : 20 / 35
Processing epoch : 21 / 35
Processing epoch : 22 / 35
Processing epoch : 23 / 35
Processing epoch : 24 / 35
Processing epoch : 25 / 35
Processing epoch : 26 / 35
Processing epoch : 27 / 35
Processing epoch : 28 / 35
Processing epoch : 29 / 35
Processing epoch : 30 / 35
Processing epoch : 31 / 35
Processing epoch : 32 / 35
Processing epoch : 33 / 35
Processing epoch : 34 / 35
Processing epoch : 35 / 35
[done]
Not setting metadata
35 matching events found
No baseline correction applied
Created an SSP operator (subspace dimension = 1)
1 projection items activated
Preparing the inverse operator for use...
Scaled noise and source covariance from nave = 1 to nave = 1
Created the regularized inverter
Created an SSP operator (subspace dimension = 1)
Created the whitener using a noise covariance matrix with rank 62 (2 small eigenvalues omitted)
Picked 64 channels from the data
Computing inverse...
Eigenleads need to be weighted ...
Processing epoch : 1 / 35
Processing epoch : 2 / 35
Processing epoch : 3 / 35
Processing epoch : 4 / 35
Processing epoch : 5 / 35
Processing epoch : 6 / 35
Processing epoch : 7 / 35
Processing epoch : 8 / 35
Processing epoch : 9 / 35
Processing epoch : 10 / 35
Processing epoch : 11 / 35
Processing epoch : 12 / 35
Processing epoch : 13 / 35
Processing epoch : 14 / 35
Processing epoch : 15 / 35
Processing epoch : 16 / 35
Processing epoch : 17 / 35
Processing epoch : 18 / 35
Processing epoch : 19 / 35
Processing epoch : 20 / 35
Processing epoch : 21 / 35
Processing epoch : 22 / 35
Processing epoch : 23 / 35
Processing epoch : 24 / 35
Processing epoch : 25 / 35
Processing epoch : 26 / 35
Processing epoch : 27 / 35
Processing epoch : 28 / 35
Processing epoch : 29 / 35
Processing epoch : 30 / 35
Processing epoch : 31 / 35
Processing epoch : 32 / 35
Processing epoch : 33 / 35
Processing epoch : 34 / 35
Processing epoch : 35 / 35
[done]
Not setting metadata
34 matching events found
No baseline correction applied
Created an SSP operator (subspace dimension = 1)
1 projection items activated
Preparing the inverse operator for use...
Scaled noise and source covariance from nave = 1 to nave = 1
Created the regularized inverter
Created an SSP operator (subspace dimension = 1)
Created the whitener using a noise covariance matrix with rank 59 (5 small eigenvalues omitted)
Picked 64 channels from the data
Computing inverse...
Eigenleads need to be weighted ...
Processing epoch : 1 / 34
Processing epoch : 2 / 34
Processing epoch : 3 / 34
Processing epoch : 4 / 34
Processing epoch : 5 / 34
Processing epoch : 6 / 34
Processing epoch : 7 / 34
Processing epoch : 8 / 34
Processing epoch : 9 / 34
Processing epoch : 10 / 34
Processing epoch : 11 / 34
Processing epoch : 12 / 34
Processing epoch : 13 / 34
Processing epoch : 14 / 34
Processing epoch : 15 / 34
Processing epoch : 16 / 34
Processing epoch : 17 / 34
Processing epoch : 18 / 34
Processing epoch : 19 / 34
Processing epoch : 20 / 34
Processing epoch : 21 / 34
Processing epoch : 22 / 34
Processing epoch : 23 / 34
Processing epoch : 24 / 34
Processing epoch : 25 / 34
Processing epoch : 26 / 34
Processing epoch : 27 / 34
Processing epoch : 28 / 34
Processing epoch : 29 / 34
Processing epoch : 30 / 34
Processing epoch : 31 / 34
Processing epoch : 32 / 34
Processing epoch : 33 / 34
Processing epoch : 34 / 34
[done]
Not setting metadata
34 matching events found
No baseline correction applied
Created an SSP operator (subspace dimension = 1)
1 projection items activated
Preparing the inverse operator for use...
Scaled noise and source covariance from nave = 1 to nave = 1
Created the regularized inverter
Created an SSP operator (subspace dimension = 1)
Created the whitener using a noise covariance matrix with rank 59 (5 small eigenvalues omitted)
Picked 64 channels from the data
Computing inverse...
Eigenleads need to be weighted ...
Processing epoch : 1 / 34
Processing epoch : 2 / 34
Processing epoch : 3 / 34
Processing epoch : 4 / 34
Processing epoch : 5 / 34
Processing epoch : 6 / 34
Processing epoch : 7 / 34
Processing epoch : 8 / 34
Processing epoch : 9 / 34
Processing epoch : 10 / 34
Processing epoch : 11 / 34
Processing epoch : 12 / 34
Processing epoch : 13 / 34
Processing epoch : 14 / 34
Processing epoch : 15 / 34
Processing epoch : 16 / 34
Processing epoch : 17 / 34
Processing epoch : 18 / 34
Processing epoch : 19 / 34
Processing epoch : 20 / 34
Processing epoch : 21 / 34
Processing epoch : 22 / 34
Processing epoch : 23 / 34
Processing epoch : 24 / 34
Processing epoch : 25 / 34
Processing epoch : 26 / 34
Processing epoch : 27 / 34
Processing epoch : 28 / 34
Processing epoch : 29 / 34
Processing epoch : 30 / 34
Processing epoch : 31 / 34
Processing epoch : 32 / 34
Processing epoch : 33 / 34
Processing epoch : 34 / 34
[done]
Not setting metadata
34 matching events found
No baseline correction applied
Created an SSP operator (subspace dimension = 1)
1 projection items activated
Preparing the inverse operator for use...
Scaled noise and source covariance from nave = 1 to nave = 1
Created the regularized inverter
Created an SSP operator (subspace dimension = 1)
Created the whitener using a noise covariance matrix with rank 59 (5 small eigenvalues omitted)
Picked 64 channels from the data
Computing inverse...
Eigenleads need to be weighted ...
Processing epoch : 1 / 34
Processing epoch : 2 / 34
Processing epoch : 3 / 34
Processing epoch : 4 / 34
Processing epoch : 5 / 34
Processing epoch : 6 / 34
Processing epoch : 7 / 34
Processing epoch : 8 / 34
Processing epoch : 9 / 34
Processing epoch : 10 / 34
Processing epoch : 11 / 34
Processing epoch : 12 / 34
Processing epoch : 13 / 34
Processing epoch : 14 / 34
Processing epoch : 15 / 34
Processing epoch : 16 / 34
Processing epoch : 17 / 34
Processing epoch : 18 / 34
Processing epoch : 19 / 34
Processing epoch : 20 / 34
Processing epoch : 21 / 34
Processing epoch : 22 / 34
Processing epoch : 23 / 34
Processing epoch : 24 / 34
Processing epoch : 25 / 34
Processing epoch : 26 / 34
Processing epoch : 27 / 34
Processing epoch : 28 / 34
Processing epoch : 29 / 34
Processing epoch : 30 / 34
Processing epoch : 31 / 34
Processing epoch : 32 / 34
Processing epoch : 33 / 34
Processing epoch : 34 / 34
[done]
Not setting metadata
34 matching events found
No baseline correction applied
Created an SSP operator (subspace dimension = 1)
1 projection items activated
Preparing the inverse operator for use...
Scaled noise and source covariance from nave = 1 to nave = 1
Created the regularized inverter
Created an SSP operator (subspace dimension = 1)
Created the whitener using a noise covariance matrix with rank 59 (5 small eigenvalues omitted)
Picked 64 channels from the data
Computing inverse...
Eigenleads need to be weighted ...
Processing epoch : 1 / 34
Processing epoch : 2 / 34
Processing epoch : 3 / 34
Processing epoch : 4 / 34
Processing epoch : 5 / 34
Processing epoch : 6 / 34
Processing epoch : 7 / 34
Processing epoch : 8 / 34
Processing epoch : 9 / 34
Processing epoch : 10 / 34
Processing epoch : 11 / 34
Processing epoch : 12 / 34
Processing epoch : 13 / 34
Processing epoch : 14 / 34
Processing epoch : 15 / 34
Processing epoch : 16 / 34
Processing epoch : 17 / 34
Processing epoch : 18 / 34
Processing epoch : 19 / 34
Processing epoch : 20 / 34
Processing epoch : 21 / 34
Processing epoch : 22 / 34
Processing epoch : 23 / 34
Processing epoch : 24 / 34
Processing epoch : 25 / 34
Processing epoch : 26 / 34
Processing epoch : 27 / 34
Processing epoch : 28 / 34
Processing epoch : 29 / 34
Processing epoch : 30 / 34
Processing epoch : 31 / 34
Processing epoch : 32 / 34
Processing epoch : 33 / 34
Processing epoch : 34 / 34
[done]
Not setting metadata
34 matching events found
No baseline correction applied
Created an SSP operator (subspace dimension = 1)
1 projection items activated
Preparing the inverse operator for use...
Scaled noise and source covariance from nave = 1 to nave = 1
Created the regularized inverter
Created an SSP operator (subspace dimension = 1)
Created the whitener using a noise covariance matrix with rank 59 (5 small eigenvalues omitted)
Picked 64 channels from the data
Computing inverse...
Eigenleads need to be weighted ...
Processing epoch : 1 / 34
Processing epoch : 2 / 34
Processing epoch : 3 / 34
Processing epoch : 4 / 34
Processing epoch : 5 / 34
Processing epoch : 6 / 34
Processing epoch : 7 / 34
Processing epoch : 8 / 34
Processing epoch : 9 / 34
Processing epoch : 10 / 34
Processing epoch : 11 / 34
Processing epoch : 12 / 34
Processing epoch : 13 / 34
Processing epoch : 14 / 34
Processing epoch : 15 / 34
Processing epoch : 16 / 34
Processing epoch : 17 / 34
Processing epoch : 18 / 34
Processing epoch : 19 / 34
Processing epoch : 20 / 34
Processing epoch : 21 / 34
Processing epoch : 22 / 34
Processing epoch : 23 / 34
Processing epoch : 24 / 34
Processing epoch : 25 / 34
Processing epoch : 26 / 34
Processing epoch : 27 / 34
Processing epoch : 28 / 34
Processing epoch : 29 / 34
Processing epoch : 30 / 34
Processing epoch : 31 / 34
Processing epoch : 32 / 34
Processing epoch : 33 / 34
Processing epoch : 34 / 34
[done]
Not setting metadata
34 matching events found
No baseline correction applied
Created an SSP operator (subspace dimension = 1)
1 projection items activated
Preparing the inverse operator for use...
Scaled noise and source covariance from nave = 1 to nave = 1
Created the regularized inverter
Created an SSP operator (subspace dimension = 1)
Created the whitener using a noise covariance matrix with rank 59 (5 small eigenvalues omitted)
Picked 64 channels from the data
Computing inverse...
Eigenleads need to be weighted ...
Processing epoch : 1 / 34
Processing epoch : 2 / 34
Processing epoch : 3 / 34
Processing epoch : 4 / 34
Processing epoch : 5 / 34
Processing epoch : 6 / 34
Processing epoch : 7 / 34
Processing epoch : 8 / 34
Processing epoch : 9 / 34
Processing epoch : 10 / 34
Processing epoch : 11 / 34
Processing epoch : 12 / 34
Processing epoch : 13 / 34
Processing epoch : 14 / 34
Processing epoch : 15 / 34
Processing epoch : 16 / 34
Processing epoch : 17 / 34
Processing epoch : 18 / 34
Processing epoch : 19 / 34
Processing epoch : 20 / 34
Processing epoch : 21 / 34
Processing epoch : 22 / 34
Processing epoch : 23 / 34
Processing epoch : 24 / 34
Processing epoch : 25 / 34
Processing epoch : 26 / 34
Processing epoch : 27 / 34
Processing epoch : 28 / 34
Processing epoch : 29 / 34
Processing epoch : 30 / 34
Processing epoch : 31 / 34
Processing epoch : 32 / 34
Processing epoch : 33 / 34
Processing epoch : 34 / 34
[done]
Not setting metadata
34 matching events found
No baseline correction applied
Created an SSP operator (subspace dimension = 1)
1 projection items activated
Preparing the inverse operator for use...
Scaled noise and source covariance from nave = 1 to nave = 1
Created the regularized inverter
Created an SSP operator (subspace dimension = 1)
Created the whitener using a noise covariance matrix with rank 59 (5 small eigenvalues omitted)
Picked 64 channels from the data
Computing inverse...
Eigenleads need to be weighted ...
Processing epoch : 1 / 34
Processing epoch : 2 / 34
Processing epoch : 3 / 34
Processing epoch : 4 / 34
Processing epoch : 5 / 34
Processing epoch : 6 / 34
Processing epoch : 7 / 34
Processing epoch : 8 / 34
Processing epoch : 9 / 34
Processing epoch : 10 / 34
Processing epoch : 11 / 34
Processing epoch : 12 / 34
Processing epoch : 13 / 34
Processing epoch : 14 / 34
Processing epoch : 15 / 34
Processing epoch : 16 / 34
Processing epoch : 17 / 34
Processing epoch : 18 / 34
Processing epoch : 19 / 34
Processing epoch : 20 / 34
Processing epoch : 21 / 34
Processing epoch : 22 / 34
Processing epoch : 23 / 34
Processing epoch : 24 / 34
Processing epoch : 25 / 34
Processing epoch : 26 / 34
Processing epoch : 27 / 34
Processing epoch : 28 / 34
Processing epoch : 29 / 34
Processing epoch : 30 / 34
Processing epoch : 31 / 34
Processing epoch : 32 / 34
Processing epoch : 33 / 34
Processing epoch : 34 / 34
[done]
Not setting metadata
34 matching events found
No baseline correction applied
Created an SSP operator (subspace dimension = 1)
1 projection items activated
Preparing the inverse operator for use...
Scaled noise and source covariance from nave = 1 to nave = 1
Created the regularized inverter
Created an SSP operator (subspace dimension = 1)
Created the whitener using a noise covariance matrix with rank 59 (5 small eigenvalues omitted)
Picked 64 channels from the data
Computing inverse...
Eigenleads need to be weighted ...
Processing epoch : 1 / 34
Processing epoch : 2 / 34
Processing epoch : 3 / 34
Processing epoch : 4 / 34
Processing epoch : 5 / 34
Processing epoch : 6 / 34
Processing epoch : 7 / 34
Processing epoch : 8 / 34
Processing epoch : 9 / 34
Processing epoch : 10 / 34
Processing epoch : 11 / 34
Processing epoch : 12 / 34
Processing epoch : 13 / 34
Processing epoch : 14 / 34
Processing epoch : 15 / 34
Processing epoch : 16 / 34
Processing epoch : 17 / 34
Processing epoch : 18 / 34
Processing epoch : 19 / 34
Processing epoch : 20 / 34
Processing epoch : 21 / 34
Processing epoch : 22 / 34
Processing epoch : 23 / 34
Processing epoch : 24 / 34
Processing epoch : 25 / 34
Processing epoch : 26 / 34
Processing epoch : 27 / 34
Processing epoch : 28 / 34
Processing epoch : 29 / 34
Processing epoch : 30 / 34
Processing epoch : 31 / 34
Processing epoch : 32 / 34
Processing epoch : 33 / 34
Processing epoch : 34 / 34
[done]
Not setting metadata
34 matching events found
No baseline correction applied
Created an SSP operator (subspace dimension = 1)
1 projection items activated
Preparing the inverse operator for use...
Scaled noise and source covariance from nave = 1 to nave = 1
Created the regularized inverter
Created an SSP operator (subspace dimension = 1)
Created the whitener using a noise covariance matrix with rank 59 (5 small eigenvalues omitted)
Picked 64 channels from the data
Computing inverse...
Eigenleads need to be weighted ...
Processing epoch : 1 / 34
Processing epoch : 2 / 34
Processing epoch : 3 / 34
Processing epoch : 4 / 34
Processing epoch : 5 / 34
Processing epoch : 6 / 34
Processing epoch : 7 / 34
Processing epoch : 8 / 34
Processing epoch : 9 / 34
Processing epoch : 10 / 34
Processing epoch : 11 / 34
Processing epoch : 12 / 34
Processing epoch : 13 / 34
Processing epoch : 14 / 34
Processing epoch : 15 / 34
Processing epoch : 16 / 34
Processing epoch : 17 / 34
Processing epoch : 18 / 34
Processing epoch : 19 / 34
Processing epoch : 20 / 34
Processing epoch : 21 / 34
Processing epoch : 22 / 34
Processing epoch : 23 / 34
Processing epoch : 24 / 34
Processing epoch : 25 / 34
Processing epoch : 26 / 34
Processing epoch : 27 / 34
Processing epoch : 28 / 34
Processing epoch : 29 / 34
Processing epoch : 30 / 34
Processing epoch : 31 / 34
Processing epoch : 32 / 34
Processing epoch : 33 / 34
Processing epoch : 34 / 34
[done]
Not setting metadata
45 matching events found
No baseline correction applied
Created an SSP operator (subspace dimension = 1)
1 projection items activated
Preparing the inverse operator for use...
Scaled noise and source covariance from nave = 1 to nave = 1
Created the regularized inverter
Created an SSP operator (subspace dimension = 1)
Created the whitener using a noise covariance matrix with rank 57 (7 small eigenvalues omitted)
Picked 64 channels from the data
Computing inverse...
Eigenleads need to be weighted ...
Processing epoch : 1 / 45
Processing epoch : 2 / 45
Processing epoch : 3 / 45
Processing epoch : 4 / 45
Processing epoch : 5 / 45
Processing epoch : 6 / 45
Processing epoch : 7 / 45
Processing epoch : 8 / 45
Processing epoch : 9 / 45
Processing epoch : 10 / 45
Processing epoch : 11 / 45
Processing epoch : 12 / 45
Processing epoch : 13 / 45
Processing epoch : 14 / 45
Processing epoch : 15 / 45
Processing epoch : 16 / 45
Processing epoch : 17 / 45
Processing epoch : 18 / 45
Processing epoch : 19 / 45
Processing epoch : 20 / 45
Processing epoch : 21 / 45
Processing epoch : 22 / 45
Processing epoch : 23 / 45
Processing epoch : 24 / 45
Processing epoch : 25 / 45
Processing epoch : 26 / 45
Processing epoch : 27 / 45
Processing epoch : 28 / 45
Processing epoch : 29 / 45
Processing epoch : 30 / 45
Processing epoch : 31 / 45
Processing epoch : 32 / 45
Processing epoch : 33 / 45
Processing epoch : 34 / 45
Processing epoch : 35 / 45
Processing epoch : 36 / 45
Processing epoch : 37 / 45
Processing epoch : 38 / 45
Processing epoch : 39 / 45
Processing epoch : 40 / 45
Processing epoch : 41 / 45
Processing epoch : 42 / 45
Processing epoch : 43 / 45
Processing epoch : 44 / 45
Processing epoch : 45 / 45
[done]
Not setting metadata
45 matching events found
No baseline correction applied
Created an SSP operator (subspace dimension = 1)
1 projection items activated
Preparing the inverse operator for use...
Scaled noise and source covariance from nave = 1 to nave = 1
Created the regularized inverter
Created an SSP operator (subspace dimension = 1)
Created the whitener using a noise covariance matrix with rank 57 (7 small eigenvalues omitted)
Picked 64 channels from the data
Computing inverse...
Eigenleads need to be weighted ...
Processing epoch : 1 / 45
Processing epoch : 2 / 45
Processing epoch : 3 / 45
Processing epoch : 4 / 45
Processing epoch : 5 / 45
Processing epoch : 6 / 45
Processing epoch : 7 / 45
Processing epoch : 8 / 45
Processing epoch : 9 / 45
Processing epoch : 10 / 45
Processing epoch : 11 / 45
Processing epoch : 12 / 45
Processing epoch : 13 / 45
Processing epoch : 14 / 45
Processing epoch : 15 / 45
Processing epoch : 16 / 45
Processing epoch : 17 / 45
Processing epoch : 18 / 45
Processing epoch : 19 / 45
Processing epoch : 20 / 45
Processing epoch : 21 / 45
Processing epoch : 22 / 45
Processing epoch : 23 / 45
Processing epoch : 24 / 45
Processing epoch : 25 / 45
Processing epoch : 26 / 45
Processing epoch : 27 / 45
Processing epoch : 28 / 45
Processing epoch : 29 / 45
Processing epoch : 30 / 45
Processing epoch : 31 / 45
Processing epoch : 32 / 45
Processing epoch : 33 / 45
Processing epoch : 34 / 45
Processing epoch : 35 / 45
Processing epoch : 36 / 45
Processing epoch : 37 / 45
Processing epoch : 38 / 45
Processing epoch : 39 / 45
Processing epoch : 40 / 45
Processing epoch : 41 / 45
Processing epoch : 42 / 45
Processing epoch : 43 / 45
Processing epoch : 44 / 45
Processing epoch : 45 / 45
[done]
Not setting metadata
45 matching events found
No baseline correction applied
Created an SSP operator (subspace dimension = 1)
1 projection items activated
Preparing the inverse operator for use...
Scaled noise and source covariance from nave = 1 to nave = 1
Created the regularized inverter
Created an SSP operator (subspace dimension = 1)
Created the whitener using a noise covariance matrix with rank 57 (7 small eigenvalues omitted)
Picked 64 channels from the data
Computing inverse...
Eigenleads need to be weighted ...
Processing epoch : 1 / 45
Processing epoch : 2 / 45
Processing epoch : 3 / 45
Processing epoch : 4 / 45
Processing epoch : 5 / 45
Processing epoch : 6 / 45
Processing epoch : 7 / 45
Processing epoch : 8 / 45
Processing epoch : 9 / 45
Processing epoch : 10 / 45
Processing epoch : 11 / 45
Processing epoch : 12 / 45
Processing epoch : 13 / 45
Processing epoch : 14 / 45
Processing epoch : 15 / 45
Processing epoch : 16 / 45
Processing epoch : 17 / 45
Processing epoch : 18 / 45
Processing epoch : 19 / 45
Processing epoch : 20 / 45
Processing epoch : 21 / 45
Processing epoch : 22 / 45
Processing epoch : 23 / 45
Processing epoch : 24 / 45
Processing epoch : 25 / 45
Processing epoch : 26 / 45
Processing epoch : 27 / 45
Processing epoch : 28 / 45
Processing epoch : 29 / 45
Processing epoch : 30 / 45
Processing epoch : 31 / 45
Processing epoch : 32 / 45
Processing epoch : 33 / 45
Processing epoch : 34 / 45
Processing epoch : 35 / 45
Processing epoch : 36 / 45
Processing epoch : 37 / 45
Processing epoch : 38 / 45
Processing epoch : 39 / 45
Processing epoch : 40 / 45
Processing epoch : 41 / 45
Processing epoch : 42 / 45
Processing epoch : 43 / 45
Processing epoch : 44 / 45
Processing epoch : 45 / 45
[done]
Not setting metadata
45 matching events found
No baseline correction applied
Created an SSP operator (subspace dimension = 1)
1 projection items activated
Preparing the inverse operator for use...
Scaled noise and source covariance from nave = 1 to nave = 1
Created the regularized inverter
Created an SSP operator (subspace dimension = 1)
Created the whitener using a noise covariance matrix with rank 57 (7 small eigenvalues omitted)
Picked 64 channels from the data
Computing inverse...
Eigenleads need to be weighted ...
Processing epoch : 1 / 45
Processing epoch : 2 / 45
Processing epoch : 3 / 45
Processing epoch : 4 / 45
Processing epoch : 5 / 45
Processing epoch : 6 / 45
Processing epoch : 7 / 45
Processing epoch : 8 / 45
Processing epoch : 9 / 45
Processing epoch : 10 / 45
Processing epoch : 11 / 45
Processing epoch : 12 / 45
Processing epoch : 13 / 45
Processing epoch : 14 / 45
Processing epoch : 15 / 45
Processing epoch : 16 / 45
Processing epoch : 17 / 45
Processing epoch : 18 / 45
Processing epoch : 19 / 45
Processing epoch : 20 / 45
Processing epoch : 21 / 45
Processing epoch : 22 / 45
Processing epoch : 23 / 45
Processing epoch : 24 / 45
Processing epoch : 25 / 45
Processing epoch : 26 / 45
Processing epoch : 27 / 45
Processing epoch : 28 / 45
Processing epoch : 29 / 45
Processing epoch : 30 / 45
Processing epoch : 31 / 45
Processing epoch : 32 / 45
Processing epoch : 33 / 45
Processing epoch : 34 / 45
Processing epoch : 35 / 45
Processing epoch : 36 / 45
Processing epoch : 37 / 45
Processing epoch : 38 / 45
Processing epoch : 39 / 45
Processing epoch : 40 / 45
Processing epoch : 41 / 45
Processing epoch : 42 / 45
Processing epoch : 43 / 45
Processing epoch : 44 / 45
Processing epoch : 45 / 45
[done]
Not setting metadata
45 matching events found
No baseline correction applied
Created an SSP operator (subspace dimension = 1)
1 projection items activated
Preparing the inverse operator for use...
Scaled noise and source covariance from nave = 1 to nave = 1
Created the regularized inverter
Created an SSP operator (subspace dimension = 1)
Created the whitener using a noise covariance matrix with rank 57 (7 small eigenvalues omitted)
Picked 64 channels from the data
Computing inverse...
Eigenleads need to be weighted ...
Processing epoch : 1 / 45
Processing epoch : 2 / 45
Processing epoch : 3 / 45
Processing epoch : 4 / 45
Processing epoch : 5 / 45
Processing epoch : 6 / 45
Processing epoch : 7 / 45
Processing epoch : 8 / 45
Processing epoch : 9 / 45
Processing epoch : 10 / 45
Processing epoch : 11 / 45
Processing epoch : 12 / 45
Processing epoch : 13 / 45
Processing epoch : 14 / 45
Processing epoch : 15 / 45
Processing epoch : 16 / 45
Processing epoch : 17 / 45
Processing epoch : 18 / 45
Processing epoch : 19 / 45
Processing epoch : 20 / 45
Processing epoch : 21 / 45
Processing epoch : 22 / 45
Processing epoch : 23 / 45
Processing epoch : 24 / 45
Processing epoch : 25 / 45
Processing epoch : 26 / 45
Processing epoch : 27 / 45
Processing epoch : 28 / 45
Processing epoch : 29 / 45
Processing epoch : 30 / 45
Processing epoch : 31 / 45
Processing epoch : 32 / 45
Processing epoch : 33 / 45
Processing epoch : 34 / 45
Processing epoch : 35 / 45
Processing epoch : 36 / 45
Processing epoch : 37 / 45
Processing epoch : 38 / 45
Processing epoch : 39 / 45
Processing epoch : 40 / 45
Processing epoch : 41 / 45
Processing epoch : 42 / 45
Processing epoch : 43 / 45
Processing epoch : 44 / 45
Processing epoch : 45 / 45
[done]
Not setting metadata
45 matching events found
No baseline correction applied
Created an SSP operator (subspace dimension = 1)
1 projection items activated
Preparing the inverse operator for use...
Scaled noise and source covariance from nave = 1 to nave = 1
Created the regularized inverter
Created an SSP operator (subspace dimension = 1)
Created the whitener using a noise covariance matrix with rank 57 (7 small eigenvalues omitted)
Picked 64 channels from the data
Computing inverse...
Eigenleads need to be weighted ...
Processing epoch : 1 / 45
Processing epoch : 2 / 45
Processing epoch : 3 / 45
Processing epoch : 4 / 45
Processing epoch : 5 / 45
Processing epoch : 6 / 45
Processing epoch : 7 / 45
Processing epoch : 8 / 45
Processing epoch : 9 / 45
Processing epoch : 10 / 45
Processing epoch : 11 / 45
Processing epoch : 12 / 45
Processing epoch : 13 / 45
Processing epoch : 14 / 45
Processing epoch : 15 / 45
Processing epoch : 16 / 45
Processing epoch : 17 / 45
Processing epoch : 18 / 45
Processing epoch : 19 / 45
Processing epoch : 20 / 45
Processing epoch : 21 / 45
Processing epoch : 22 / 45
Processing epoch : 23 / 45
Processing epoch : 24 / 45
Processing epoch : 25 / 45
Processing epoch : 26 / 45
Processing epoch : 27 / 45
Processing epoch : 28 / 45
Processing epoch : 29 / 45
Processing epoch : 30 / 45
Processing epoch : 31 / 45
Processing epoch : 32 / 45
Processing epoch : 33 / 45
Processing epoch : 34 / 45
Processing epoch : 35 / 45
Processing epoch : 36 / 45
Processing epoch : 37 / 45
Processing epoch : 38 / 45
Processing epoch : 39 / 45
Processing epoch : 40 / 45
Processing epoch : 41 / 45
Processing epoch : 42 / 45
Processing epoch : 43 / 45
Processing epoch : 44 / 45
Processing epoch : 45 / 45
[done]
Not setting metadata
45 matching events found
No baseline correction applied
Created an SSP operator (subspace dimension = 1)
1 projection items activated
Preparing the inverse operator for use...
Scaled noise and source covariance from nave = 1 to nave = 1
Created the regularized inverter
Created an SSP operator (subspace dimension = 1)
Created the whitener using a noise covariance matrix with rank 57 (7 small eigenvalues omitted)
Picked 64 channels from the data
Computing inverse...
Eigenleads need to be weighted ...
Processing epoch : 1 / 45
Processing epoch : 2 / 45
Processing epoch : 3 / 45
Processing epoch : 4 / 45
Processing epoch : 5 / 45
Processing epoch : 6 / 45
Processing epoch : 7 / 45
Processing epoch : 8 / 45
Processing epoch : 9 / 45
Processing epoch : 10 / 45
Processing epoch : 11 / 45
Processing epoch : 12 / 45
Processing epoch : 13 / 45
Processing epoch : 14 / 45
Processing epoch : 15 / 45
Processing epoch : 16 / 45
Processing epoch : 17 / 45
Processing epoch : 18 / 45
Processing epoch : 19 / 45
Processing epoch : 20 / 45
Processing epoch : 21 / 45
Processing epoch : 22 / 45
Processing epoch : 23 / 45
Processing epoch : 24 / 45
Processing epoch : 25 / 45
Processing epoch : 26 / 45
Processing epoch : 27 / 45
Processing epoch : 28 / 45
Processing epoch : 29 / 45
Processing epoch : 30 / 45
Processing epoch : 31 / 45
Processing epoch : 32 / 45
Processing epoch : 33 / 45
Processing epoch : 34 / 45
Processing epoch : 35 / 45
Processing epoch : 36 / 45
Processing epoch : 37 / 45
Processing epoch : 38 / 45
Processing epoch : 39 / 45
Processing epoch : 40 / 45
Processing epoch : 41 / 45
Processing epoch : 42 / 45
Processing epoch : 43 / 45
Processing epoch : 44 / 45
Processing epoch : 45 / 45
[done]
Not setting metadata
45 matching events found
No baseline correction applied
Created an SSP operator (subspace dimension = 1)
1 projection items activated
Preparing the inverse operator for use...
Scaled noise and source covariance from nave = 1 to nave = 1
Created the regularized inverter
Created an SSP operator (subspace dimension = 1)
Created the whitener using a noise covariance matrix with rank 57 (7 small eigenvalues omitted)
Picked 64 channels from the data
Computing inverse...
Eigenleads need to be weighted ...
Processing epoch : 1 / 45
Processing epoch : 2 / 45
Processing epoch : 3 / 45
Processing epoch : 4 / 45
Processing epoch : 5 / 45
Processing epoch : 6 / 45
Processing epoch : 7 / 45
Processing epoch : 8 / 45
Processing epoch : 9 / 45
Processing epoch : 10 / 45
Processing epoch : 11 / 45
Processing epoch : 12 / 45
Processing epoch : 13 / 45
Processing epoch : 14 / 45
Processing epoch : 15 / 45
Processing epoch : 16 / 45
Processing epoch : 17 / 45
Processing epoch : 18 / 45
Processing epoch : 19 / 45
Processing epoch : 20 / 45
Processing epoch : 21 / 45
Processing epoch : 22 / 45
Processing epoch : 23 / 45
Processing epoch : 24 / 45
Processing epoch : 25 / 45
Processing epoch : 26 / 45
Processing epoch : 27 / 45
Processing epoch : 28 / 45
Processing epoch : 29 / 45
Processing epoch : 30 / 45
Processing epoch : 31 / 45
Processing epoch : 32 / 45
Processing epoch : 33 / 45
Processing epoch : 34 / 45
Processing epoch : 35 / 45
Processing epoch : 36 / 45
Processing epoch : 37 / 45
Processing epoch : 38 / 45
Processing epoch : 39 / 45
Processing epoch : 40 / 45
Processing epoch : 41 / 45
Processing epoch : 42 / 45
Processing epoch : 43 / 45
Processing epoch : 44 / 45
Processing epoch : 45 / 45
[done]
Not setting metadata
45 matching events found
No baseline correction applied
Created an SSP operator (subspace dimension = 1)
1 projection items activated
Preparing the inverse operator for use...
Scaled noise and source covariance from nave = 1 to nave = 1
Created the regularized inverter
Created an SSP operator (subspace dimension = 1)
Created the whitener using a noise covariance matrix with rank 57 (7 small eigenvalues omitted)
Picked 64 channels from the data
Computing inverse...
Eigenleads need to be weighted ...
Processing epoch : 1 / 45
Processing epoch : 2 / 45
Processing epoch : 3 / 45
Processing epoch : 4 / 45
Processing epoch : 5 / 45
Processing epoch : 6 / 45
Processing epoch : 7 / 45
Processing epoch : 8 / 45
Processing epoch : 9 / 45
Processing epoch : 10 / 45
Processing epoch : 11 / 45
Processing epoch : 12 / 45
Processing epoch : 13 / 45
Processing epoch : 14 / 45
Processing epoch : 15 / 45
Processing epoch : 16 / 45
Processing epoch : 17 / 45
Processing epoch : 18 / 45
Processing epoch : 19 / 45
Processing epoch : 20 / 45
Processing epoch : 21 / 45
Processing epoch : 22 / 45
Processing epoch : 23 / 45
Processing epoch : 24 / 45
Processing epoch : 25 / 45
Processing epoch : 26 / 45
Processing epoch : 27 / 45
Processing epoch : 28 / 45
Processing epoch : 29 / 45
Processing epoch : 30 / 45
Processing epoch : 31 / 45
Processing epoch : 32 / 45
Processing epoch : 33 / 45
Processing epoch : 34 / 45
Processing epoch : 35 / 45
Processing epoch : 36 / 45
Processing epoch : 37 / 45
Processing epoch : 38 / 45
Processing epoch : 39 / 45
Processing epoch : 40 / 45
Processing epoch : 41 / 45
Processing epoch : 42 / 45
Processing epoch : 43 / 45
Processing epoch : 44 / 45
Processing epoch : 45 / 45
[done]
Traceback (most recent call last):
File "/home/circleci/python_env/lib/python3.10/site-packages/matplotlib/cbook.py", line 361, in process
func(*args, **kwargs)
File "/home/circleci/python_env/lib/python3.10/site-packages/matplotlib/colorbar.py", line 513, in update_normal
self._draw_all()
File "/home/circleci/python_env/lib/python3.10/site-packages/matplotlib/colorbar.py", line 570, in _draw_all
self._add_solids(X, Y, self._values[ind, np.newaxis])
File "/home/circleci/python_env/lib/python3.10/site-packages/matplotlib/colorbar.py", line 576, in _add_solids
self.solids.remove()
File "/home/circleci/python_env/lib/python3.10/site-packages/matplotlib/artist.py", line 256, in remove
raise NotImplementedError('cannot remove artist')
NotImplementedError: cannot remove artist
Traceback (most recent call last):
File "/home/circleci/python_env/lib/python3.10/site-packages/matplotlib/cbook.py", line 361, in process
func(*args, **kwargs)
File "/home/circleci/python_env/lib/python3.10/site-packages/matplotlib/colorbar.py", line 513, in update_normal
self._draw_all()
File "/home/circleci/python_env/lib/python3.10/site-packages/matplotlib/colorbar.py", line 570, in _draw_all
self._add_solids(X, Y, self._values[ind, np.newaxis])
File "/home/circleci/python_env/lib/python3.10/site-packages/matplotlib/colorbar.py", line 576, in _add_solids
self.solids.remove()
File "/home/circleci/python_env/lib/python3.10/site-packages/matplotlib/artist.py", line 256, in remove
raise NotImplementedError('cannot remove artist')
NotImplementedError: cannot remove artist
Total running time of the script: (6 minutes 53.629 seconds)
Estimated memory usage: 4330 MB