Note
Go to the end to download the full example code or to run this example in your browser via Binder.
Utilising Auxiliary Data#
In this example we demonstrate how to load auxiliary data from a SNIRF file and include it in the design matrix for incorporating with your GLM analysis.
This example builds on the GLM tutorial. As such, we will not explain the GLM procedure in this example and refer readers to the detailed description above. Instead, we focus on extracting the auxiliary data and how this can be incorporated in to your analysis.
# sphinx_gallery_thumbnail_number = 2
# Authors: Robert Luke <code@robertluke.net>
#
# License: BSD (3-clause)
import matplotlib.pyplot as plt
import mne
import numpy as np
import pandas as pd
from nilearn.plotting import plot_design_matrix
from mne_nirs.channels import get_long_channels, get_short_channels
from mne_nirs.datasets.snirf_with_aux import data_path
from mne_nirs.experimental_design import make_first_level_design_matrix
from mne_nirs.io.snirf import read_snirf_aux_data
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
Import raw NIRS data#
First we import the raw data. A different dataset is used from the previous GLM example that contains auxiliary data.
fnirs_snirf_file = data_path()
raw_intensity = mne.io.read_raw_snirf(fnirs_snirf_file).load_data()
raw_intensity.resample(0.7)
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
Clean up annotations before analysis#
Next we update the annotations by assigning names to each trigger ID. Then we crop the recording to the section containing our experimental conditions.
raw_intensity.annotations.rename(
{"1": "Control", "2": "Tapping_Left", "3": "Tapping_Right"}
)
raw_intensity.annotations.delete(raw_intensity.annotations.description == "15")
raw_intensity.annotations.set_durations(5)
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
<Annotations | 60 segments: Control (20), Tapping_Left (20), Tapping_Right ...>
Preprocess NIRS data#
Next we convert the raw data to haemoglobin concentration.
raw_od = mne.preprocessing.nirs.optical_density(raw_intensity)
raw_haemo = mne.preprocessing.nirs.beer_lambert_law(raw_od, ppf=0.1)
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
We then split the data in to short channels which predominantly contain systemic responses and long channels which have both neural and systemic contributions.
short_chs = get_short_channels(raw_haemo)
raw_haemo = get_long_channels(raw_haemo)
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
Create design matrix#
Next we create a model to fit our data to. The model consists of various components to model different things we assume contribute to the measured signal.
design_matrix = make_first_level_design_matrix(
raw_haemo,
drift_model="cosine",
high_pass=0.005, # Must be specified per experiment
hrf_model="spm",
stim_dur=5.0,
)
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
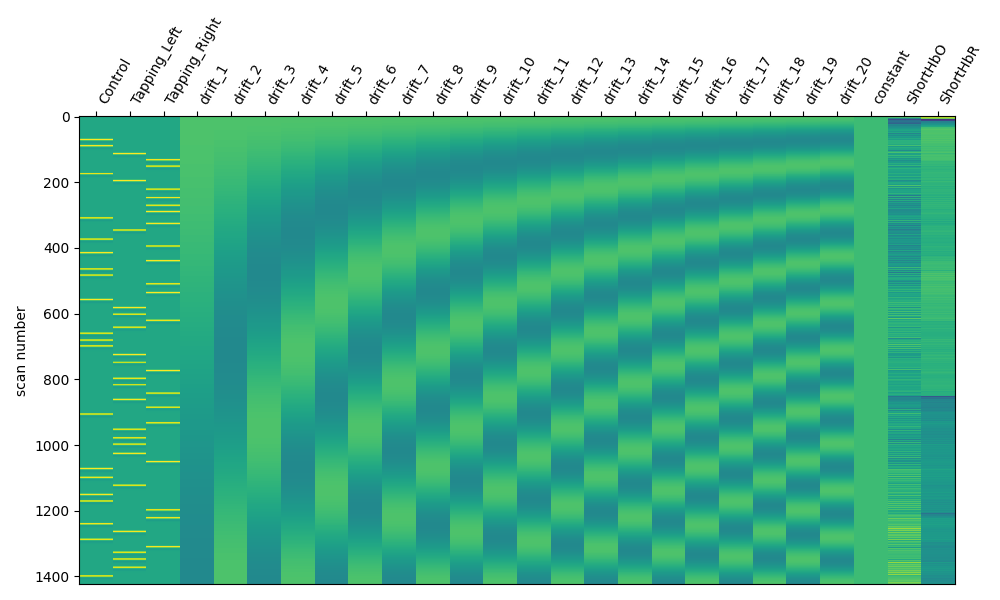
We also add the mean of the short channels to the design matrix. In theory these channels contain only systemic components, so including them in the design matrix allows us to estimate the neural component related to each experimental condition uncontaminated by systemic effects.
design_matrix["ShortHbO"] = np.mean(
short_chs.copy().pick(picks="hbo").get_data(), axis=0
)
design_matrix["ShortHbR"] = np.mean(
short_chs.copy().pick(picks="hbr").get_data(), axis=0
)
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
We can view the design matrix by printing the variable and we see that it includes the standard regressors, but does not yet contain any auxiliary data.
fig, ax1 = plt.subplots(figsize=(10, 6), nrows=1, ncols=1)
fig = plot_design_matrix(design_matrix, axes=ax1)
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
Load auxiliary data#
The design matrix is a pandas data frame. As such, we wish to load the auxiliary data in the same format. The following function will load the SNIRF file and extract the auxiliary data. The auxiliary data can be sampled at a different rate to the raw fNIRS data, so this function will conveniently resample the data to the same rate as the raw fNIRS data.
aux_df = read_snirf_aux_data(fnirs_snirf_file, raw_haemo)
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
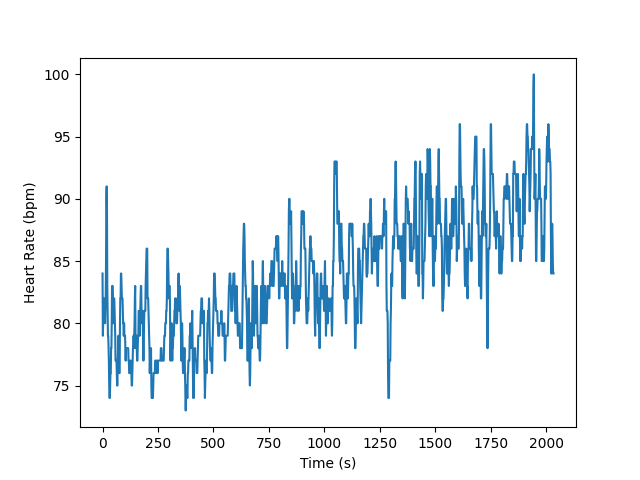
And you can verify the data looks reasonable by plotting individual fields.
plt.plot(raw_haemo.times, aux_df["HR"])
plt.xlabel("Time (s)")
plt.ylabel("Heart Rate (bpm)")
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
Text(38.347222222222214, 0.5, 'Heart Rate (bpm)')
Include auxiliary data in design matrix#
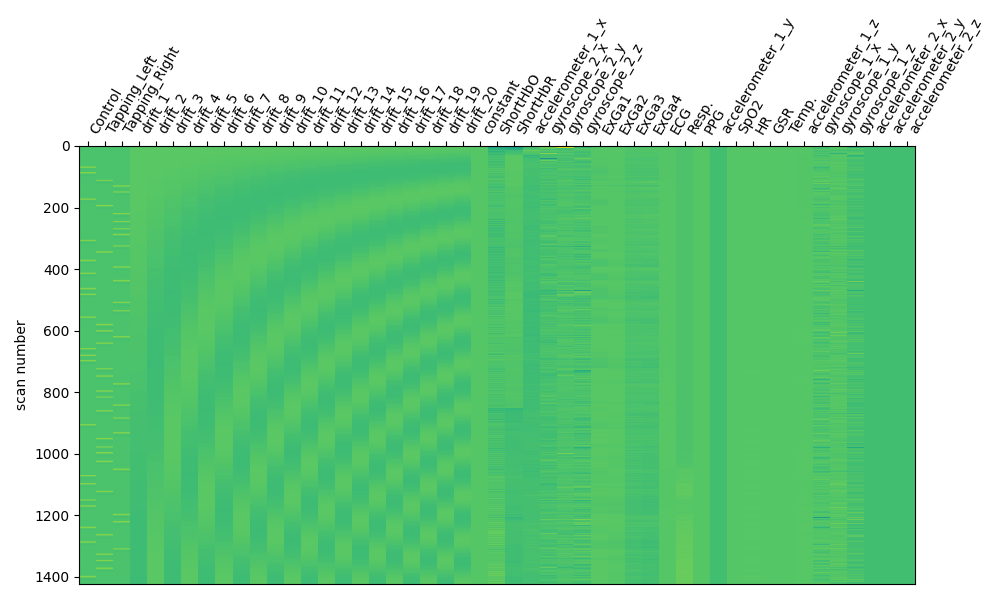
Finally we append the auxiliary data to the design matrix so that these can be included as regressors in a GLM analysis.
design_matrix = pd.concat([design_matrix, aux_df], axis=1)
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
And we can visually display the design matrix and verify the data is included.
fig, ax1 = plt.subplots(figsize=(10, 6), nrows=1, ncols=1)
fig = plot_design_matrix(design_matrix, axes=ax1)
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
/usr/lib/python3.12/multiprocessing/popen_fork.py:66: DeprecationWarning: This process (pid=7130) is multi-threaded, use of fork() may lead to deadlocks in the child.
self.pid = os.fork()
Conclusion#
We have demonstrated how to load auxiliary data from a SNIRF file. We illustrated how to include this data in your design matrix for further GLM analysis. We do not go through a full GLM analysis, instead the reader is directed to the dedicated GLM tutorial. The auxiliary data may need to be treated before being included in your analysis, for example you may need to normalise before inclusion in statistical analysis etc, but this is beyond the scope of this tutorial.
Total running time of the script: (0 minutes 6.098 seconds)
Estimated memory usage: 510 MB