Note
Go to the end to download the full example code or to run this example in your browser via Binder.
Signal Enhancement Techniques#
In this document we investigate the effect of different signal enhancement techniques on functional near-infrared spectroscopy (fNIRS) data.
# Authors: Robert Luke <mail@robertluke.net>
#
# License: BSD (3-clause)
import os
import matplotlib.pyplot as plt
import mne
import mne_nirs
from mne_nirs.channels import picks_pair_to_idx
Import and preprocess data#
This code is exactly the same as the first sections in the MNE tutorial. See https://mne.tools/dev/auto_tutorials/preprocessing/plot_70_fnirs_processing.html for more details.
fnirs_data_folder = mne.datasets.fnirs_motor.data_path()
fnirs_raw_dir = os.path.join(fnirs_data_folder, "Participant-1")
raw_intensity = mne.io.read_raw_nirx(fnirs_raw_dir, verbose=True).load_data()
raw_od = mne.preprocessing.nirs.optical_density(raw_intensity)
raw_haemo = mne.preprocessing.nirs.beer_lambert_law(raw_od, ppf=0.1)
raw_haemo = mne_nirs.channels.get_long_channels(raw_haemo)
raw_haemo = raw_haemo.filter(0.05, 0.7, h_trans_bandwidth=0.2, l_trans_bandwidth=0.02)
events, _ = mne.events_from_annotations(
raw_haemo, event_id={"1.0": 1, "2.0": 2, "3.0": 3}
)
event_dict = {"Control": 1, "Tapping/Left": 2, "Tapping/Right": 3}
Loading /home/circleci/mne_data/MNE-fNIRS-motor-data/Participant-1
Extract epochs with no additional processing#
First we extract the epochs with no additional processing, this result should be the same as the MNE tutorial.
reject_criteria = dict(hbo=100e-6)
tmin, tmax = -5, 15
epochs = mne.Epochs(
raw_haemo,
events,
event_id=event_dict,
tmin=tmin,
tmax=tmax,
reject=reject_criteria,
reject_by_annotation=True,
proj=True,
baseline=(None, 0),
preload=True,
detrend=None,
verbose=True,
)
evoked_dict = {
"Tapping/HbO": epochs["Tapping"].average(picks="hbo"),
"Tapping/HbR": epochs["Tapping"].average(picks="hbr"),
"Control/HbO": epochs["Control"].average(picks="hbo"),
"Control/HbR": epochs["Control"].average(picks="hbr"),
}
# Rename channels until the encoding of frequency in ch_name is fixed
for condition in evoked_dict:
evoked_dict[condition].rename_channels(lambda x: x[:-4])
Not setting metadata
90 matching events found
Setting baseline interval to [-4.992, 0.0] s
Applying baseline correction (mode: mean)
0 projection items activated
Using data from preloaded Raw for 90 events and 157 original time points ...
Rejecting epoch based on HBO : ['S4_D4 hbo']
Rejecting epoch based on HBO : ['S4_D4 hbo']
Rejecting epoch based on HBO : ['S4_D4 hbo', 'S8_D8 hbo']
Rejecting epoch based on HBO : ['S8_D8 hbo']
Rejecting epoch based on HBO : ['S4_D4 hbo']
5 bad epochs dropped
Apply negative correlation enhancement algorithm#
Apply Cui et. al. 2010 and extract epochs.
raw_anti = mne_nirs.signal_enhancement.enhance_negative_correlation(raw_haemo)
epochs_anti = mne.Epochs(
raw_anti,
events,
event_id=event_dict,
tmin=tmin,
tmax=tmax,
reject=reject_criteria,
reject_by_annotation=True,
proj=True,
baseline=(None, 0),
preload=True,
detrend=None,
verbose=True,
)
evoked_dict_anti = {
"Tapping/HbO": epochs_anti["Tapping"].average(picks="hbo"),
"Tapping/HbR": epochs_anti["Tapping"].average(picks="hbr"),
"Control/HbO": epochs_anti["Control"].average(picks="hbo"),
"Control/HbR": epochs_anti["Control"].average(picks="hbr"),
}
# Rename channels until the encoding of frequency in ch_name is fixed
for condition in evoked_dict_anti:
evoked_dict_anti[condition].rename_channels(lambda x: x[:-4])
Not setting metadata
90 matching events found
Setting baseline interval to [-4.992, 0.0] s
Applying baseline correction (mode: mean)
0 projection items activated
Using data from preloaded Raw for 90 events and 157 original time points ...
0 bad epochs dropped
Apply short channel correction#
Apply Scholkmann et al 2014 and extract epochs.
od_corrected = mne_nirs.signal_enhancement.short_channel_regression(raw_od)
raw_haemo = mne.preprocessing.nirs.beer_lambert_law(od_corrected, ppf=0.1)
raw_haemo = mne_nirs.channels.get_long_channels(raw_haemo)
raw_haemo = raw_haemo.filter(0.05, 0.7, h_trans_bandwidth=0.2, l_trans_bandwidth=0.02)
epochs_corr = mne.Epochs(
raw_haemo,
events,
event_id=event_dict,
tmin=tmin,
tmax=tmax,
reject=reject_criteria,
reject_by_annotation=True,
proj=True,
baseline=(None, 0),
preload=True,
detrend=None,
verbose=True,
)
evoked_dict_corr = {
"Tapping/HbO": epochs_corr["Tapping"].average(picks="hbo"),
"Tapping/HbR": epochs_corr["Tapping"].average(picks="hbr"),
"Control/HbO": epochs_corr["Control"].average(picks="hbo"),
"Control/HbR": epochs_corr["Control"].average(picks="hbr"),
}
# Rename channels until the encoding of frequency in ch_name is fixed
for condition in evoked_dict_corr:
evoked_dict_corr[condition].rename_channels(lambda x: x[:-4])
Not setting metadata
90 matching events found
Setting baseline interval to [-4.992, 0.0] s
Applying baseline correction (mode: mean)
0 projection items activated
Using data from preloaded Raw for 90 events and 157 original time points ...
Rejecting epoch based on HBO : ['S4_D4 hbo', 'S8_D8 hbo']
Rejecting epoch based on HBO : ['S1_D1 hbo', 'S7_D6 hbo', 'S7_D7 hbo', 'S8_D8 hbo']
Rejecting epoch based on HBO : ['S4_D4 hbo', 'S8_D8 hbo']
3 bad epochs dropped
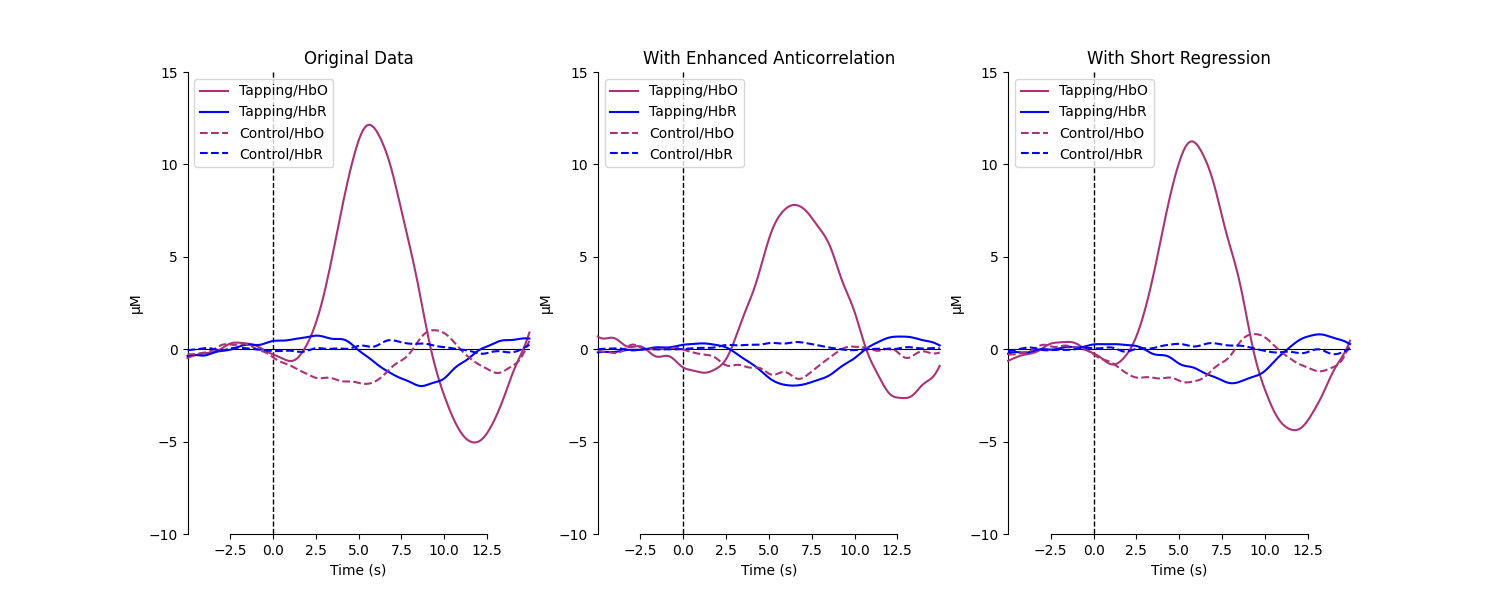
Plot approaches for comparison#
Plot the average epochs with and without Cui 2010 applied.
fig, axes = plt.subplots(nrows=1, ncols=3, figsize=(15, 6))
color_dict = dict(HbO="#AA3377", HbR="b")
styles_dict = dict(Control=dict(linestyle="dashed"))
mne.viz.plot_compare_evokeds(
evoked_dict,
combine="mean",
ci=0.95,
axes=axes[0],
colors=color_dict,
styles=styles_dict,
ylim=dict(hbo=[-10, 15]),
)
mne.viz.plot_compare_evokeds(
evoked_dict_anti,
combine="mean",
ci=0.95,
axes=axes[1],
colors=color_dict,
styles=styles_dict,
ylim=dict(hbo=[-10, 15]),
)
mne.viz.plot_compare_evokeds(
evoked_dict_corr,
combine="mean",
ci=0.95,
axes=axes[2],
colors=color_dict,
styles=styles_dict,
ylim=dict(hbo=[-10, 15]),
)
for column, condition in enumerate(
["Original Data", "With Enhanced Anticorrelation", "With Short Regression"]
):
axes[column].set_title(f"{condition}")
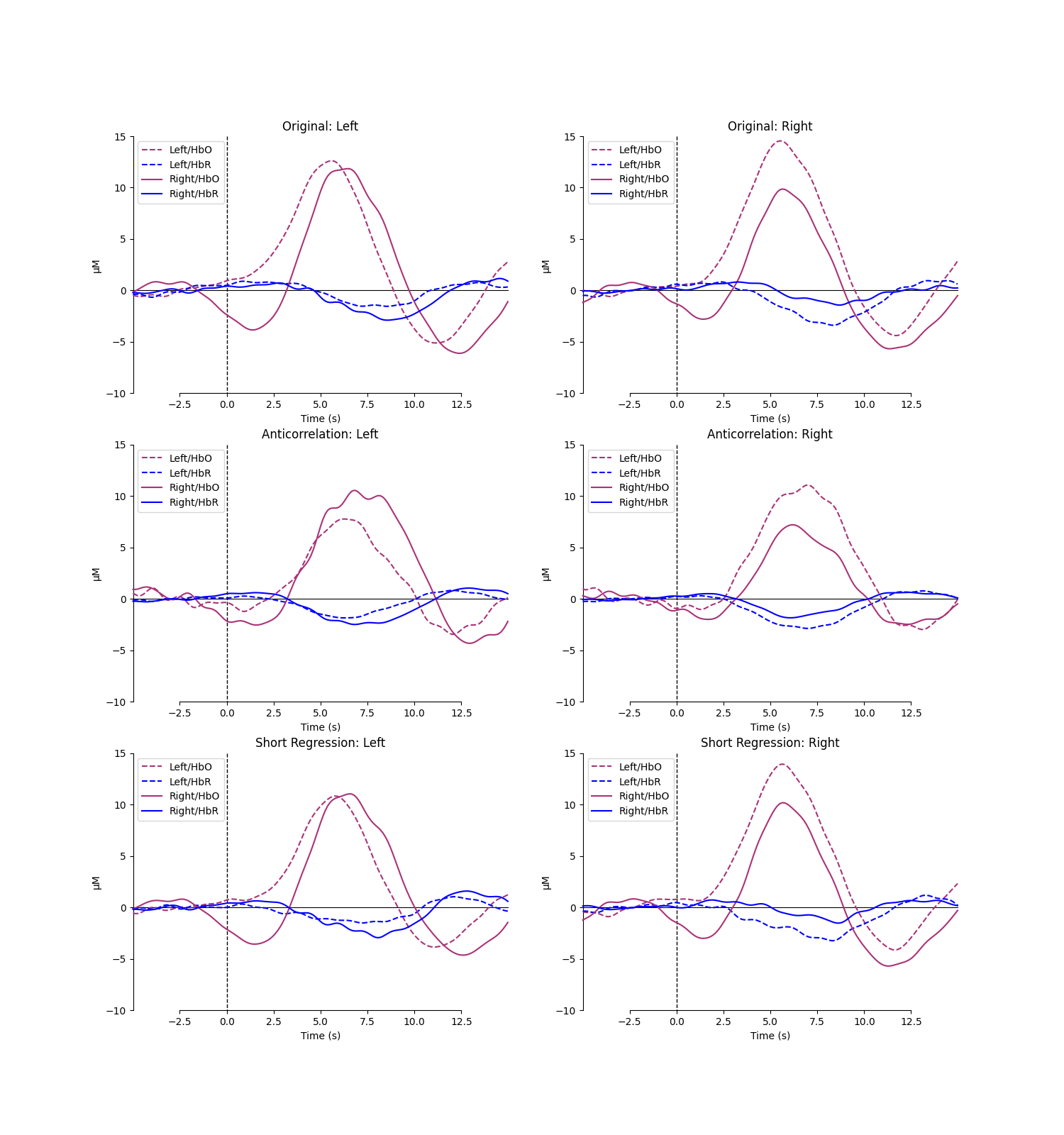
Plot hemisphere for each approach#
Plot the epoch image for each approach. First we specify the source detector pairs for analysis.
left = [[1, 3], [2, 3], [1, 2], [4, 3]]
right = [[5, 7], [6, 7], [5, 6], [8, 7]]
groups = dict(
Left_ROI=picks_pair_to_idx(raw_anti.pick(picks="hbo"), left, on_missing="warning"),
Right_ROI=picks_pair_to_idx(
raw_anti.pick(picks="hbo"), right, on_missing="warning"
),
)
evoked_dict = {
"Left/HbO": epochs["Tapping/Left"].average(picks="hbo"),
"Left/HbR": epochs["Tapping/Left"].average(picks="hbr"),
"Right/HbO": epochs["Tapping/Right"].average(picks="hbo"),
"Right/HbR": epochs["Tapping/Right"].average(picks="hbr"),
}
for condition in evoked_dict:
evoked_dict[condition].rename_channels(lambda x: x[:-4])
evoked_dict_anti = {
"Left/HbO": epochs_anti["Tapping/Left"].average(picks="hbo"),
"Left/HbR": epochs_anti["Tapping/Left"].average(picks="hbr"),
"Right/HbO": epochs_anti["Tapping/Right"].average(picks="hbo"),
"Right/HbR": epochs_anti["Tapping/Right"].average(picks="hbr"),
}
for condition in evoked_dict_anti:
evoked_dict_anti[condition].rename_channels(lambda x: x[:-4])
evoked_dict_corr = {
"Left/HbO": epochs_corr["Tapping/Left"].average(picks="hbo"),
"Left/HbR": epochs_corr["Tapping/Left"].average(picks="hbr"),
"Right/HbO": epochs_corr["Tapping/Right"].average(picks="hbo"),
"Right/HbR": epochs_corr["Tapping/Right"].average(picks="hbr"),
}
for condition in evoked_dict_corr:
evoked_dict_corr[condition].rename_channels(lambda x: x[:-4])
color_dict = dict(HbO="#AA3377", HbR="b")
styles_dict = dict(Left=dict(linestyle="dashed"))
fig, axes = plt.subplots(nrows=3, ncols=2, figsize=(15, 16))
mne.viz.plot_compare_evokeds(
evoked_dict,
combine="mean",
ci=0.95,
picks=groups["Left_ROI"],
axes=axes[0, 0],
colors=color_dict,
styles=styles_dict,
ylim=dict(hbo=[-10, 15]),
)
mne.viz.plot_compare_evokeds(
evoked_dict,
combine="mean",
ci=0.95,
picks=groups["Right_ROI"],
axes=axes[0, 1],
colors=color_dict,
styles=styles_dict,
ylim=dict(hbo=[-10, 15]),
)
mne.viz.plot_compare_evokeds(
evoked_dict_anti,
combine="mean",
ci=0.95,
picks=groups["Left_ROI"],
axes=axes[1, 0],
colors=color_dict,
styles=styles_dict,
ylim=dict(hbo=[-10, 15]),
)
mne.viz.plot_compare_evokeds(
evoked_dict_anti,
combine="mean",
ci=0.95,
picks=groups["Right_ROI"],
axes=axes[1, 1],
colors=color_dict,
styles=styles_dict,
ylim=dict(hbo=[-10, 15]),
)
mne.viz.plot_compare_evokeds(
evoked_dict_corr,
combine="mean",
ci=0.95,
picks=groups["Left_ROI"],
axes=axes[2, 0],
colors=color_dict,
styles=styles_dict,
ylim=dict(hbo=[-10, 15]),
)
mne.viz.plot_compare_evokeds(
evoked_dict_corr,
combine="mean",
ci=0.95,
picks=groups["Right_ROI"],
axes=axes[2, 1],
colors=color_dict,
styles=styles_dict,
ylim=dict(hbo=[-10, 15]),
)
for row, condition in enumerate(["Original", "Anticorrelation", "Short Regression"]):
for column, hemi in enumerate(["Left", "Right"]):
axes[row, column].set_title(f"{condition}: {hemi}")
Total running time of the script: (0 minutes 3.620 seconds)
Estimated memory usage: 511 MB