Note
Click here to download the full example code
Reading an inverse operator#
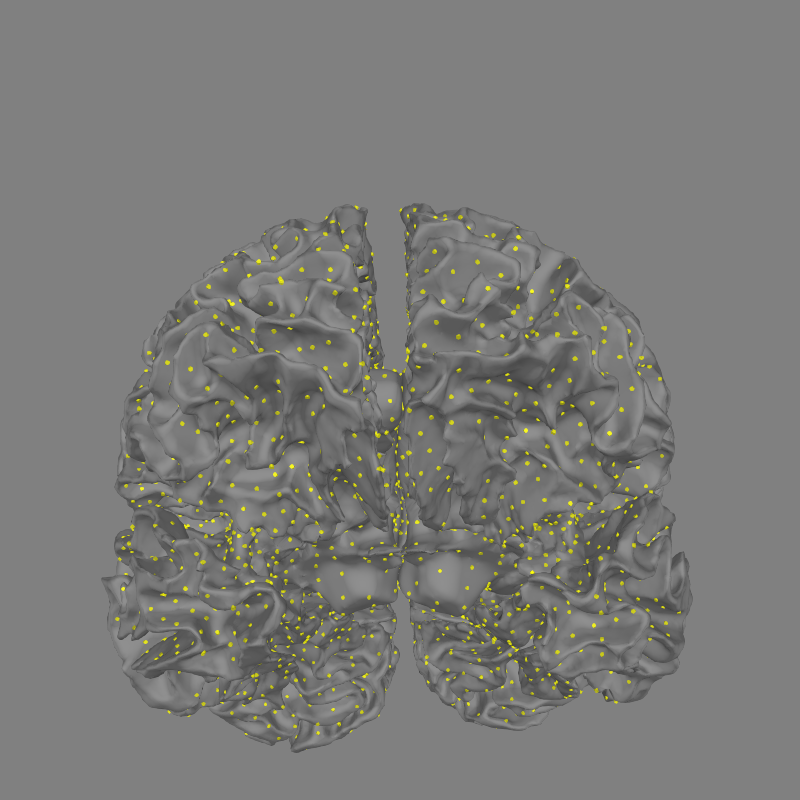
The inverse operator’s source space is shown in 3D.
# Author: Alexandre Gramfort <alexandre.gramfort@inria.fr>
#
# License: BSD-3-Clause
import mne
from mne.datasets import sample
from mne.minimum_norm import read_inverse_operator
from mne.viz import set_3d_view
print(__doc__)
data_path = sample.data_path()
subjects_dir = data_path / 'subjects'
meg_path = data_path / 'MEG' / 'sample'
fname_trans = meg_path / 'sample_audvis_raw-trans.fif'
inv_fname = meg_path / 'sample_audvis-meg-oct-6-meg-inv.fif'
inv = read_inverse_operator(inv_fname)
print("Method: %s" % inv['methods'])
print("fMRI prior: %s" % inv['fmri_prior'])
print("Number of sources: %s" % inv['nsource'])
print("Number of channels: %s" % inv['nchan'])
src = inv['src'] # get the source space
# Get access to the triangulation of the cortex
print("Number of vertices on the left hemisphere: %d" % len(src[0]['rr']))
print("Number of triangles on left hemisphere: %d" % len(src[0]['use_tris']))
print("Number of vertices on the right hemisphere: %d" % len(src[1]['rr']))
print("Number of triangles on right hemisphere: %d" % len(src[1]['use_tris']))
Reading inverse operator decomposition from /home/circleci/mne_data/MNE-sample-data/MEG/sample/sample_audvis-meg-oct-6-meg-inv.fif...
Reading inverse operator info...
[done]
Reading inverse operator decomposition...
[done]
305 x 305 full covariance (kind = 1) found.
Read a total of 4 projection items:
PCA-v1 (1 x 102) active
PCA-v2 (1 x 102) active
PCA-v3 (1 x 102) active
Average EEG reference (1 x 60) active
Noise covariance matrix read.
22494 x 22494 diagonal covariance (kind = 2) found.
Source covariance matrix read.
22494 x 22494 diagonal covariance (kind = 6) found.
Orientation priors read.
22494 x 22494 diagonal covariance (kind = 5) found.
Depth priors read.
Did not find the desired covariance matrix (kind = 3)
Reading a source space...
Computing patch statistics...
Patch information added...
Distance information added...
[done]
Reading a source space...
Computing patch statistics...
Patch information added...
Distance information added...
[done]
2 source spaces read
Read a total of 4 projection items:
PCA-v1 (1 x 102) active
PCA-v2 (1 x 102) active
PCA-v3 (1 x 102) active
Average EEG reference (1 x 60) active
Source spaces transformed to the inverse solution coordinate frame
Method: 1
fMRI prior: None
Number of sources: 7498
Number of channels: 305
Number of vertices on the left hemisphere: 155407
Number of triangles on left hemisphere: 8192
Number of vertices on the right hemisphere: 156866
Number of triangles on right hemisphere: 8192
Show the 3D source space
fig = mne.viz.plot_alignment(subject='sample', subjects_dir=subjects_dir,
trans=fname_trans, surfaces='white', src=src)
set_3d_view(fig, focalpoint=(0., 0., 0.06))
Total running time of the script: ( 0 minutes 5.219 seconds)
Estimated memory usage: 15 MB