Note
Click here to download the full example code
Working with events#
This tutorial describes event representation and how event arrays are used to subselect data.
As usual we’ll start by importing the modules we need, loading some
example data, and cropping the Raw
object to just 60 seconds before loading it into RAM to save memory:
import os
import numpy as np
import mne
sample_data_folder = mne.datasets.sample.data_path()
sample_data_raw_file = os.path.join(sample_data_folder, 'MEG', 'sample',
'sample_audvis_raw.fif')
raw = mne.io.read_raw_fif(sample_data_raw_file, verbose=False)
raw.crop(tmax=60).load_data()
Reading 0 ... 36037 = 0.000 ... 60.000 secs...
The tutorial Parsing events from raw data describes in detail the
different ways of obtaining an Events array from a
Raw object (see the section
Detecting experimental events for details). Since the sample
dataset includes experimental events recorded on
stim channel STI 014, we’ll start this tutorial by parsing the
events from that channel using mne.find_events():
events = mne.find_events(raw, stim_channel='STI 014')
86 events found
Event IDs: [ 1 2 3 4 5 32]
Reading and writing events from/to a file#
Event arrays are NumPy array objects, so they could
be saved to disk as binary .npy files using numpy.save().
However, MNE-Python provides convenience functions mne.read_events()
and mne.write_events() for reading and writing event arrays as either
text files (common file extensions are .eve, .lst, and
.txt) or binary .fif files. The example dataset includes the
results of mne.find_events(raw) in a .fif file. Since we’ve
truncated our Raw object, it will have fewer events than the
events file loaded from disk (which contains events for the entire
recording), but the events should match for the first 60 seconds anyway:
sample_data_events_file = os.path.join(sample_data_folder, 'MEG', 'sample',
'sample_audvis_raw-eve.fif')
events_from_file = mne.read_events(sample_data_events_file)
assert np.array_equal(events, events_from_file[:len(events)])
When writing event arrays to disk, the format will be inferred from the file
extension you provide. By convention, MNE-Python expects events files to
either have an .eve extension or to have a file basename ending in
-eve or _eve (e.g., my_experiment_eve.fif), and will issue
a warning if this convention is not respected.
Subselecting and combining events#
The output of find_events() above (repeated here) told us the
number of events that were found, and the unique integer event IDs present:
mne.find_events(raw, stim_channel='STI 014')
86 events found
Event IDs: [ 1 2 3 4 5 32]
If some of those events are not of interest, you can easily subselect events
using mne.pick_events(), which has parameters include and
exclude. For example, in the sample data Event ID 32 corresponds to a
subject button press, which could be excluded as:
events_no_button = mne.pick_events(events, exclude=32)
It is also possible to combine two Event IDs using mne.merge_events();
the following example will combine Event IDs 1, 2 and 3 into a single event
labelled 1:
merged_events = mne.merge_events(events, [1, 2, 3], 1)
print(np.unique(merged_events[:, -1]))
[ 1 4 5 32]
Note, however, that merging events is not necessary if you simply want to pool trial types for analysis; the next section describes how MNE-Python uses event dictionaries to map integer Event IDs to more descriptive label strings.
Mapping Event IDs to trial descriptors#
So far in this tutorial we’ve only been dealing with integer Event IDs, which were assigned based on DC voltage pulse magnitude (which is ultimately determined by the experimenter’s choices about what signals to send to the STIM channels). Keeping track of which Event ID corresponds to which experimental condition can be cumbersome, and it is often desirable to pool experimental conditions during analysis. You may recall that the mapping of integer Event IDs to meaningful descriptions for the sample dataset is given in this table in the introductory tutorial. Here we simply reproduce that mapping as an event dictionary:
event_dict = {'auditory/left': 1, 'auditory/right': 2, 'visual/left': 3,
'visual/right': 4, 'smiley': 5, 'buttonpress': 32}
Event dictionaries like this one are used when extracting epochs from
continuous data, and the resulting Epochs object allows pooling
by requesting partial trial descriptors. For example, if we wanted to pool
all auditory trials, instead of merging Event IDs 1 and 2 using the
merge_events() function, we can make use of the fact that the keys
of event_dict contain multiple trial descriptors separated by /
characters: requesting 'auditory' trials will select all epochs with
Event IDs 1 and 2; requesting 'left' trials will select all epochs with
Event IDs 1 and 3. An example of this is shown later, in the
Subselecting epochs section of the tutorial
The Epochs data structure: discontinuous data.
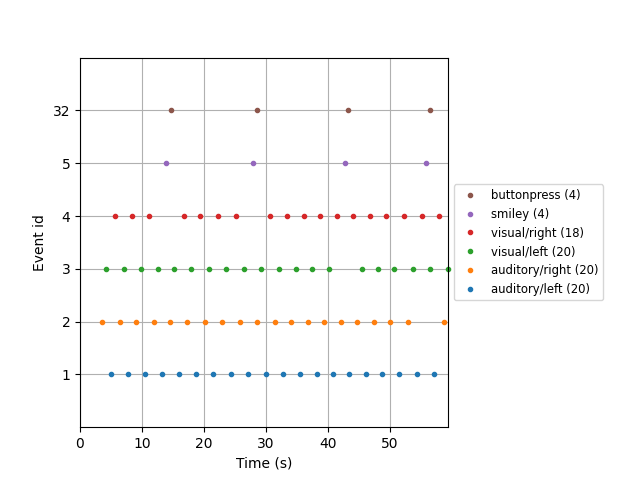
Plotting events#
Another use of event dictionaries is when plotting events, which can serve as
a useful check that your event signals were properly sent to the STIM
channel(s) and that MNE-Python has successfully found them. The function
mne.viz.plot_events() will plot each event versus its sample number
(or, if you provide the sampling frequency, it will plot them versus time in
seconds). It can also account for the offset between sample number and sample
index in Neuromag systems, with the first_samp parameter. If an event
dictionary is provided, it will be used to generate a legend:
fig = mne.viz.plot_events(events, sfreq=raw.info['sfreq'],
first_samp=raw.first_samp, event_id=event_dict)
fig.subplots_adjust(right=0.7) # make room for legend
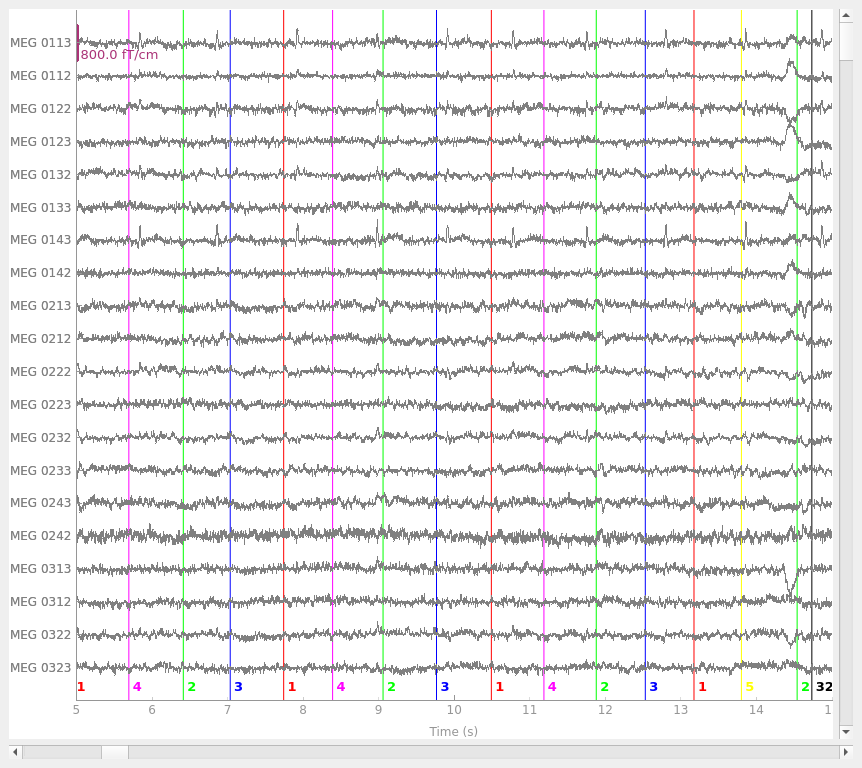
Plotting events and raw data together#
Events can also be plotted alongside the Raw object they
were extracted from, by passing the Event array as the events parameter
of raw.plot:
Making equally-spaced Events arrays#
For some experiments (such as those intending to analyze resting-state
activity) there may not be any experimental events included in the raw
recording. In such cases, an Events array of equally-spaced events can be
generated using mne.make_fixed_length_events():
new_events = mne.make_fixed_length_events(raw, start=5, stop=50, duration=2.)
By default, the events will all be given the integer Event ID of 1, but
you can change that with the id parameter. It is also possible to specify
an overlap duration — i.e., if you ultimately want epochs that
are 2.5 seconds long, but you want them to overlap by 0.5 seconds, you can
specify duration=2.5, overlap=0.5 in the call to
make_fixed_length_events() (this will yield the same spacing of
events as duration=2, overlap=0).
Total running time of the script: ( 0 minutes 5.437 seconds)
Estimated memory usage: 121 MB