Note
Click here to download the full example code
Sensitivity map of SSP projections#
This example shows the sources that have a forward field similar to the first SSP vector correcting for ECG.
# Author: Alexandre Gramfort <alexandre.gramfort@inria.fr>
#
# License: BSD-3-Clause
import matplotlib.pyplot as plt
from mne import read_forward_solution, read_proj, sensitivity_map
from mne.datasets import sample
print(__doc__)
data_path = sample.data_path()
subjects_dir = data_path / 'subjects'
meg_path = data_path / 'MEG' / 'sample'
fname = meg_path / 'sample_audvis-meg-eeg-oct-6-fwd.fif'
ecg_fname = meg_path / 'sample_audvis_ecg-proj.fif'
fwd = read_forward_solution(fname)
projs = read_proj(ecg_fname)
# take only one projection per channel type
projs = projs[::2]
# Compute sensitivity map
ssp_ecg_map = sensitivity_map(fwd, ch_type='grad', projs=projs, mode='angle')
Reading forward solution from /home/circleci/mne_data/MNE-sample-data/MEG/sample/sample_audvis-meg-eeg-oct-6-fwd.fif...
Reading a source space...
Computing patch statistics...
Patch information added...
Distance information added...
[done]
Reading a source space...
Computing patch statistics...
Patch information added...
Distance information added...
[done]
2 source spaces read
Desired named matrix (kind = 3523) not available
Read MEG forward solution (7498 sources, 306 channels, free orientations)
Desired named matrix (kind = 3523) not available
Read EEG forward solution (7498 sources, 60 channels, free orientations)
MEG and EEG forward solutions combined
Source spaces transformed to the forward solution coordinate frame
Read a total of 6 projection items:
ECG-planar-999--0.200-0.400-PCA-01 (1 x 203) idle
ECG-planar-999--0.200-0.400-PCA-02 (1 x 203) idle
ECG-axial-999--0.200-0.400-PCA-01 (1 x 102) idle
ECG-axial-999--0.200-0.400-PCA-02 (1 x 102) idle
ECG-eeg-999--0.200-0.400-PCA-01 (1 x 59) idle
ECG-eeg-999--0.200-0.400-PCA-02 (1 x 59) idle
204 out of 366 channels remain after picking
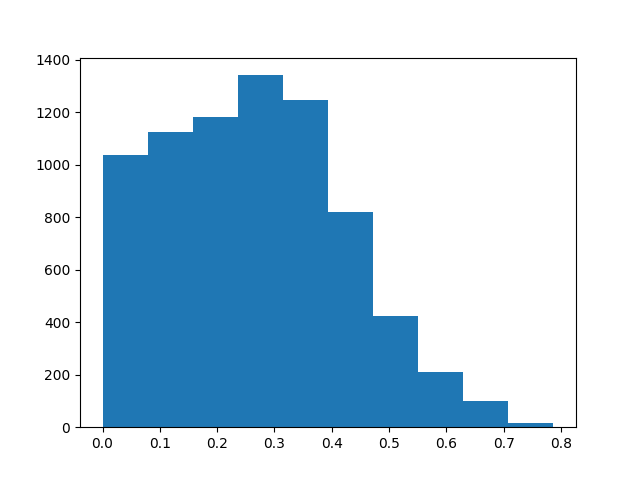
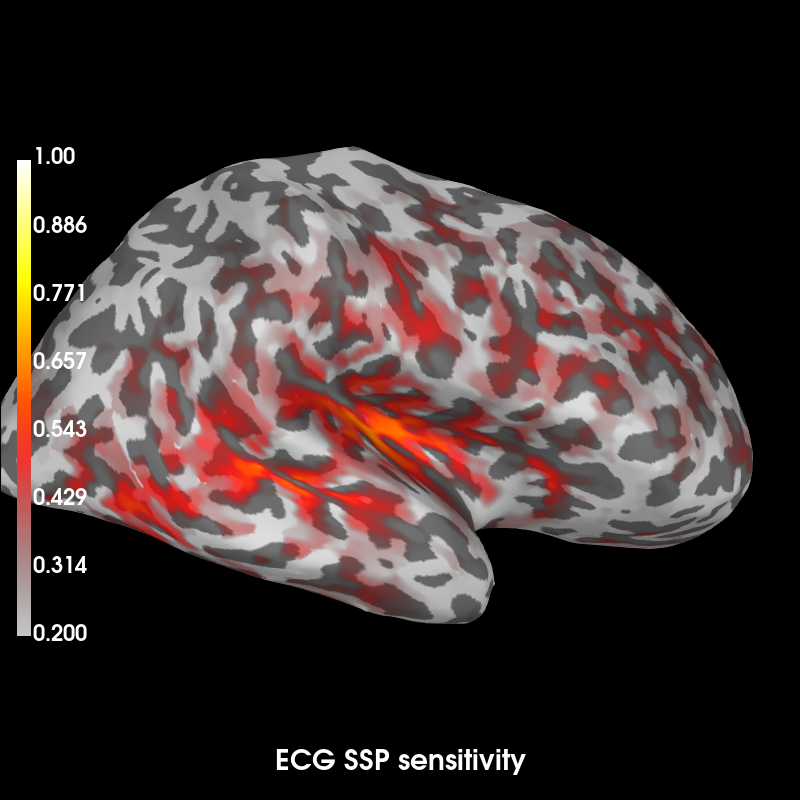
Show sensitivity map
plt.hist(ssp_ecg_map.data.ravel())
plt.show()
args = dict(clim=dict(kind='value', lims=(0.2, 0.6, 1.)), smoothing_steps=7,
hemi='rh', subjects_dir=subjects_dir)
ssp_ecg_map.plot(subject='sample', time_label='ECG SSP sensitivity', **args)
Total running time of the script: ( 0 minutes 6.758 seconds)
Estimated memory usage: 46 MB