Note
Go to the end to download the full example code
Plot a cortical parcellation#
In this example, we download the HCP-MMP1.0 parcellation
[1] and show it on fsaverage.
We will also download the customized 448-label aparc
parcellation from [2].
Note
The HCP-MMP dataset has license terms restricting its use. Of particular relevance:
“I will acknowledge the use of WU-Minn HCP data and data derived from WU-Minn HCP data when publicly presenting any results or algorithms that benefitted from their use.”
# Author: Eric Larson <larson.eric.d@gmail.com>
# Denis Engemann <denis.engemann@gmail.com>
#
# License: BSD-3-Clause
import mne
Brain = mne.viz.get_brain_class()
subjects_dir = mne.datasets.sample.data_path() / 'subjects'
mne.datasets.fetch_hcp_mmp_parcellation(subjects_dir=subjects_dir,
verbose=True)
mne.datasets.fetch_aparc_sub_parcellation(subjects_dir=subjects_dir,
verbose=True)
labels = mne.read_labels_from_annot(
'fsaverage', 'HCPMMP1', 'lh', subjects_dir=subjects_dir)
brain = Brain('fsaverage', 'lh', 'inflated', subjects_dir=subjects_dir,
cortex='low_contrast', background='white', size=(800, 600))
brain.add_annotation('HCPMMP1')
aud_label = [label for label in labels if label.name == 'L_A1_ROI-lh'][0]
brain.add_label(aud_label, borders=False)
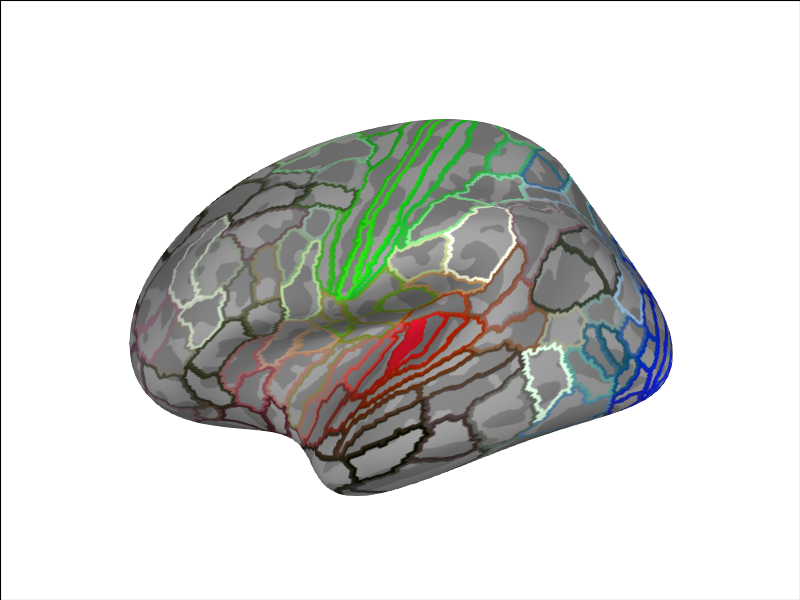
Reading labels from parcellation...
read 181 labels from /home/circleci/mne_data/MNE-sample-data/subjects/fsaverage/label/lh.HCPMMP1.annot
We can also plot a combined set of labels (23 per hemisphere).
brain = Brain('fsaverage', 'lh', 'inflated', subjects_dir=subjects_dir,
cortex='low_contrast', background='white', size=(800, 600))
brain.add_annotation('HCPMMP1_combined')
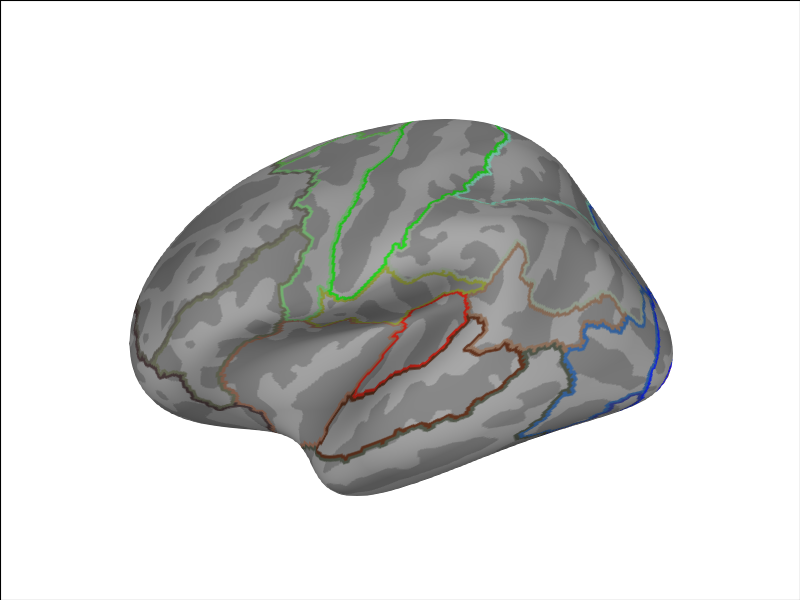
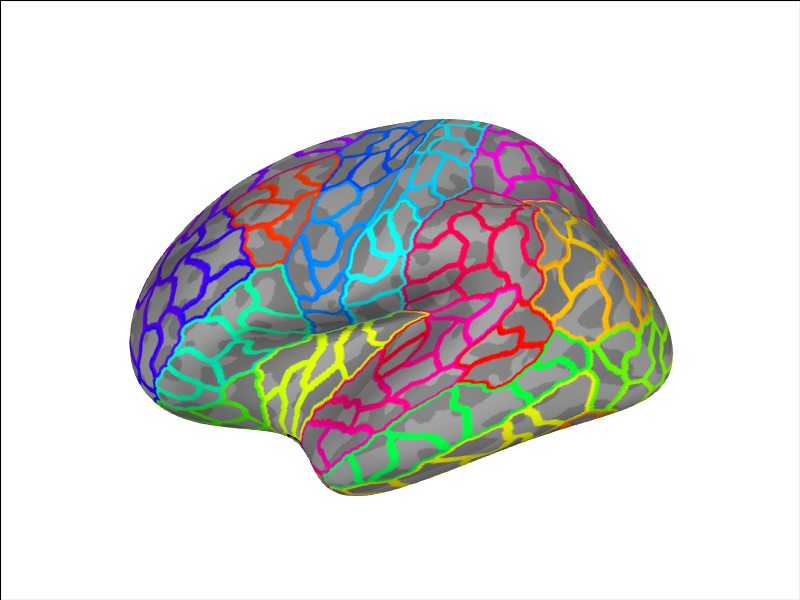
We can add another custom parcellation
brain = Brain('fsaverage', 'lh', 'inflated', subjects_dir=subjects_dir,
cortex='low_contrast', background='white', size=(800, 600))
brain.add_annotation('aparc_sub')
References#
Total running time of the script: ( 0 minutes 8.599 seconds)
Estimated memory usage: 27 MB