Note
Go to the end to download the full example code
Cross-hemisphere comparison#
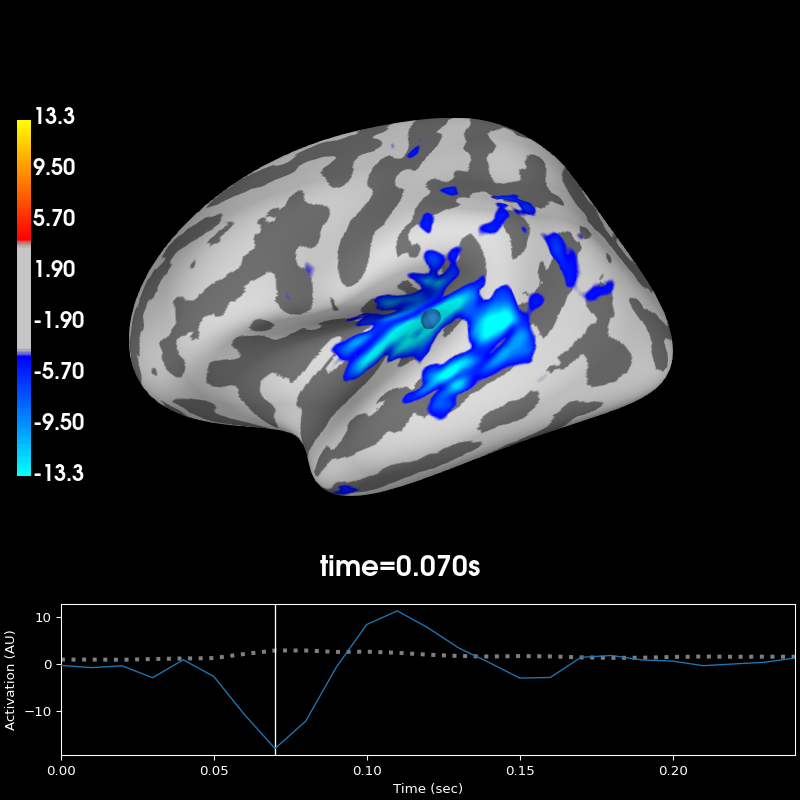
This example illustrates how to visualize the difference between activity in
the left and the right hemisphere. The data from the right hemisphere is
mapped to the left hemisphere, and then the difference is plotted. For more
information see mne.compute_source_morph().
# Author: Christian Brodbeck <christianbrodbeck@nyu.edu>
#
# License: BSD-3-Clause
import mne
data_dir = mne.datasets.sample.data_path()
subjects_dir = data_dir / 'subjects'
stc_path = data_dir / 'MEG' / 'sample' / 'sample_audvis-meg-eeg'
stc = mne.read_source_estimate(stc_path, 'sample')
# First, morph the data to fsaverage_sym, for which we have left_right
# registrations:
stc = mne.compute_source_morph(stc, 'sample', 'fsaverage_sym', smooth=5,
warn=False,
subjects_dir=subjects_dir).apply(stc)
# Compute a morph-matrix mapping the right to the left hemisphere,
# and vice-versa.
morph = mne.compute_source_morph(stc, 'fsaverage_sym', 'fsaverage_sym',
spacing=stc.vertices, warn=False,
subjects_dir=subjects_dir, xhemi=True,
verbose='error') # creating morph map
stc_xhemi = morph.apply(stc)
# Now we can subtract them and plot the result:
diff = stc - stc_xhemi
diff.plot(hemi='lh', subjects_dir=subjects_dir, initial_time=0.07,
size=(800, 600))
surface source space present ...
Computing morph matrix...
Left-hemisphere map read.
Right-hemisphere map read.
5 smooth iterations done.
5 smooth iterations done.
[done]
[done]
Using control points [ 3.70314401 4.48867635 13.29876034]
Total running time of the script: ( 0 minutes 4.610 seconds)
Estimated memory usage: 23 MB