Note
Go to the end to download the full example code
FreeSurfer MRI reconstruction#
This tutorial covers how to use FreeSurfer alongside MNE-Python, to handle the structural MRI data that we use to build subject-specific anatomical models of the scalp, inner/outer skull, and cortical surface.
FreeSurfer is an open source analysis toolbox for MRI data, available from
https://surfer.nmr.mgh.harvard.edu/. FreeSurfer provides graphical interfaces
for visualizing MRI data, several anatomical parcellations useful for creating
region-of-interest (ROI) labels, template brains such as fsaverage, and
several command-line tools for tasks like finding tissue boundaries or morphing
brains to align analogous anatomical regions across subjects.
These FreeSurfer capabilities are necessary for MNE-Python to compute the
forward model and set up the corresponding source space (a grid of dipoles located on the cortical surface or
within the brain volume).
First steps#
After downloading and installing FreeSurfer, there are a few steps to set up
the environment. First is to define an environment variable FREESURFER_HOME
and then run the FreeSurfer setup script:
$ export FREESURFER_HOME=/path/to/FreeSurfer
$ source $FREESURFER_HOME/SetUpFreeSurfer.sh
Note
The FreeSurfer home directory will vary depending on your operating system and choices you made during installation. See the FreeSurfer installation guide for more info.
Another important step is to tell FreeSurfer where to put the anatomical
reconstructions of your research subjects. This is done with an environment
variable called SUBJECTS_DIR, which will contain the individual subjects’
reconstructions in separate sub-folders.
$ export SUBJECTS_DIR=/path/to/your/subjects_dir
Again see the FreeSurfer installation guide for more info.
Anatomical reconstruction#
The first processing stage is the creation of various surface reconstructions. Usually a full FreeSurfer reconstruction is obtained by the following commands:
$ my_subject=sample
$ my_NIfTI=/path/to/NIfTI.nii.gz
$ recon-all -i $my_NIfTI -s $my_subject -all
where i stands for “input” and s for “subject”. Executing this will
create the folder $SUBJECTS_DIR/sample and populate it
with several subfolders (bem, label, mri, etc). See also the
FreeSurfer wiki’s recommended reconstruction workflow for more detailed
explanation.
Warning
Anatomical reconstruction can take several hours, even on a fast computer.
FreeSurfer performs a hemispheric separation so most resulting files have
separate left and right hemisphere versions, indicated by the prefix
lh or rh. This hemispheric separation is preserved by MNE-Python (e.g.,
mne.SourceEstimate objects store spatial locations (vertices) for the two
hemispheres separately; cf. The SourceEstimate data structure).
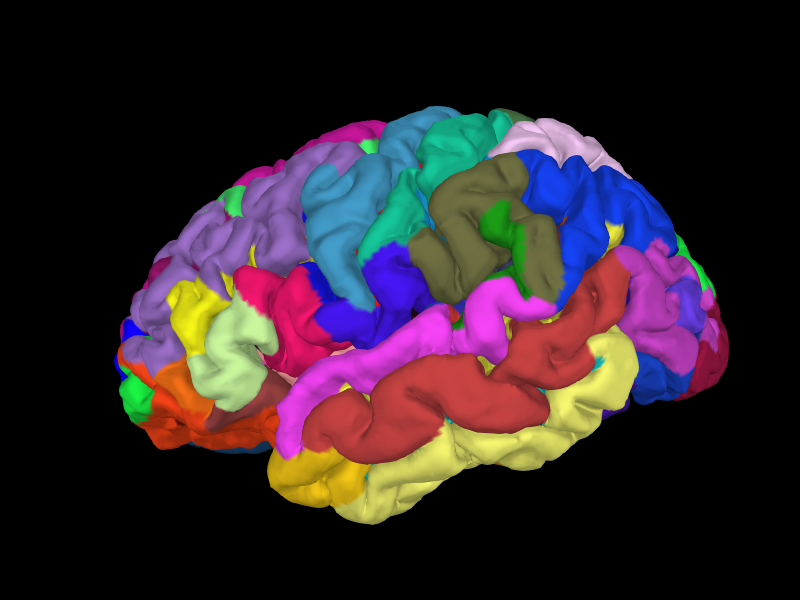
Below we show an example of the results of a FreeSurfer reconstruction for the left hemisphere of the Sample dataset subject, including an overlay of an anatomical parcellation (in this case, the parcellation from [1]).
import mne
sample_data_folder = mne.datasets.sample.data_path()
subjects_dir = sample_data_folder / 'subjects'
Brain = mne.viz.get_brain_class()
brain = Brain('sample', hemi='lh', surf='pial',
subjects_dir=subjects_dir, size=(800, 600))
brain.add_annotation('aparc.a2009s', borders=False)
Use with MNE-Python#
For source localization analysis to work properly, it is important that the
FreeSurfer reconstruction has completed beforehand. Furthermore, for many
MNE-Python functions related to inverse imaging (such as
mne.setup_source_space), SUBJECTS_DIR has to be defined globally (as an
environment variable or through a call to mne.set_config), or specified
separately in each function call by passing the keyword argument
subjects_dir='/path/to/your/subjects_dir'.
See Setting up the source space to get an idea of how this works for one particular function, and How MNE uses FreeSurfer’s outputs for more details on how MNE-Python and FreeSurfer are integrated.
‘fsaverage’#
During installation, FreeSurfer copies a subject called 'fsaverage' to
$FREESURFER_HOME/subjects/fsaverage. fsaverage is a template brain
based on a combination of 40 MRI scans of real brains. The fsaverage
subject folder contains all the files that a normal subject reconstruction
would yield. See https://surfer.nmr.mgh.harvard.edu/fswiki/FsAverage for an
overview, and https://surfer.nmr.mgh.harvard.edu/fswiki/Buckner40Notes for
details about the subjects used to create fsaverage. A copy of
fsaverage is also provided as part of the Sample dataset
and is also distributed as a standalone dataset.
One of the most common uses of fsaverage is as a destination space for
cortical morphing / source estimate transformations. In
other words, it is common to morph each individual subject’s estimated brain
activity onto the fsaverage brain, so that group-level statistical
comparisons can be made.
References#
Total running time of the script: ( 0 minutes 2.932 seconds)
Estimated memory usage: 21 MB