mne.read_source_spaces#
- mne.read_source_spaces(fname, patch_stats=False, verbose=None)[source]#
Read the source spaces from a FIF file.
- Parameters:
- fname
str The name of the file, which should end with -src.fif or -src.fif.gz.
- patch_stats
bool, optional (defaultFalse) Calculate and add cortical patch statistics to the surfaces.
- verbose
bool|str|int|None Control verbosity of the logging output. If
None, use the default verbosity level. See the logging documentation andmne.verbose()for details. Should only be passed as a keyword argument.
- fname
- Returns:
- src
SourceSpaces The source spaces.
- src
Examples using mne.read_source_spaces#
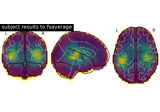
Permutation t-test on source data with spatio-temporal clustering
Permutation t-test on source data with spatio-temporal clustering
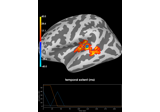
2 samples permutation test on source data with spatio-temporal clustering
2 samples permutation test on source data with spatio-temporal clustering
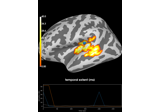
Repeated measures ANOVA on source data with spatio-temporal clustering
Repeated measures ANOVA on source data with spatio-temporal clustering

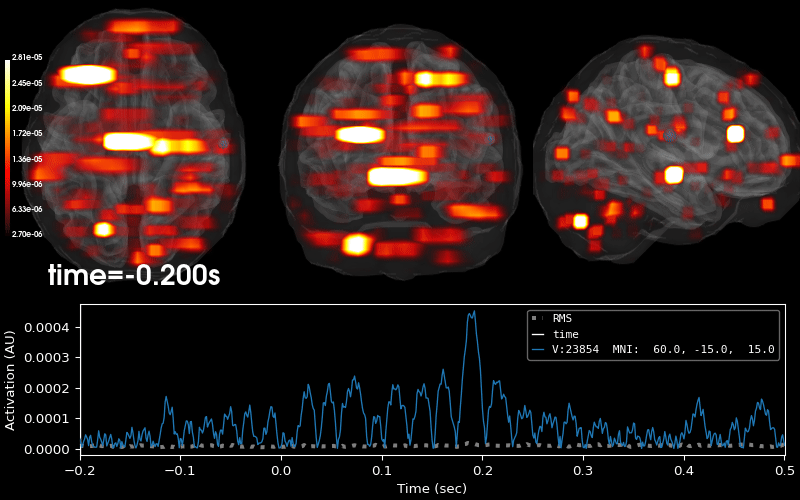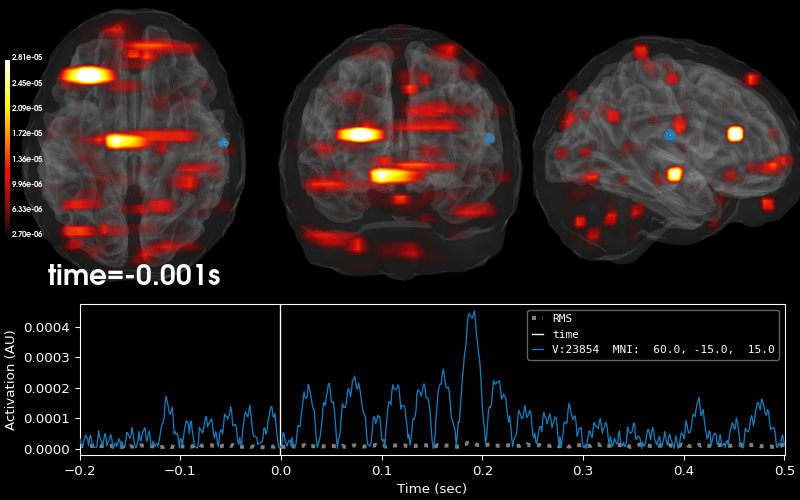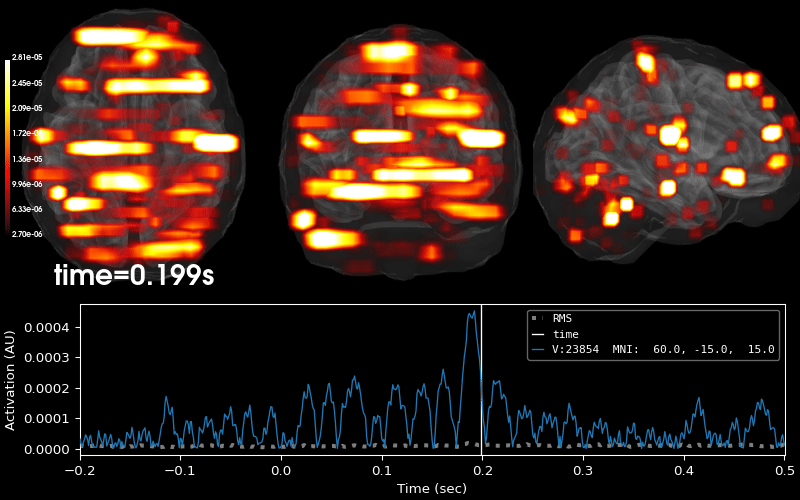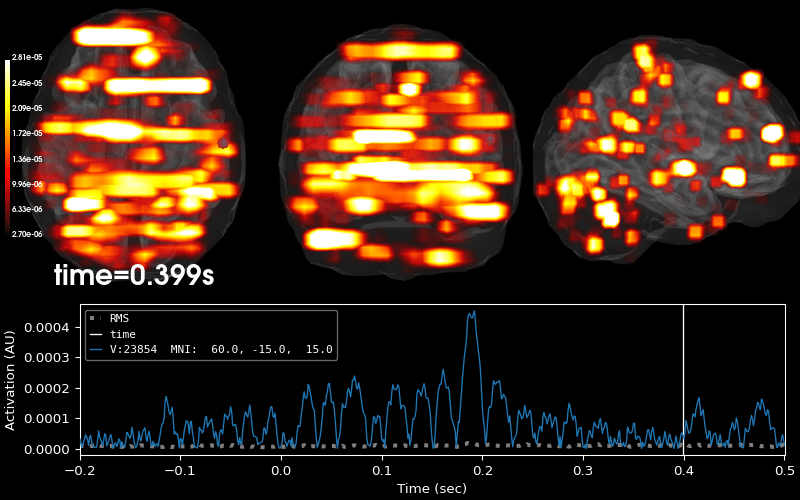
Compute sparse inverse solution with mixed norm: MxNE and irMxNE
Compute sparse inverse solution with mixed norm: MxNE and irMxNE