Note
Go to the end to download the full example code
Brainstorm Elekta phantom dataset tutorial#
Here we compute the evoked from raw for the Brainstorm Elekta phantom tutorial dataset. For comparison, see [1] and the original Brainstorm tutorial.
# Authors: Eric Larson <larson.eric.d@gmail.com>
#
# License: BSD-3-Clause
# Copyright the MNE-Python contributors.
import matplotlib.pyplot as plt
import numpy as np
import mne
from mne import find_events, fit_dipole
from mne.datasets import fetch_phantom
from mne.datasets.brainstorm import bst_phantom_elekta
from mne.io import read_raw_fif
print(__doc__)
The data were collected with an Elekta Neuromag VectorView system at 1000 Hz
and low-pass filtered at 330 Hz. Here the medium-amplitude (200 nAm) data
are read to construct instances of mne.io.Raw.
Opening raw data file /home/circleci/mne_data/MNE-brainstorm-data/bst_phantom_elekta/kojak_all_200nAm_pp_no_chpi_no_ms_raw.fif...
Read a total of 13 projection items:
planar-0.0-115.0-PCA-01 (1 x 306) idle
planar-0.0-115.0-PCA-02 (1 x 306) idle
planar-0.0-115.0-PCA-03 (1 x 306) idle
planar-0.0-115.0-PCA-04 (1 x 306) idle
planar-0.0-115.0-PCA-05 (1 x 306) idle
axial-0.0-115.0-PCA-01 (1 x 306) idle
axial-0.0-115.0-PCA-02 (1 x 306) idle
axial-0.0-115.0-PCA-03 (1 x 306) idle
axial-0.0-115.0-PCA-04 (1 x 306) idle
axial-0.0-115.0-PCA-05 (1 x 306) idle
axial-0.0-115.0-PCA-06 (1 x 306) idle
axial-0.0-115.0-PCA-07 (1 x 306) idle
axial-0.0-115.0-PCA-08 (1 x 306) idle
Range : 47000 ... 437999 = 47.000 ... 437.999 secs
Ready.
Data channel array consisted of 204 MEG planor gradiometers, 102 axial magnetometers, and 3 stimulus channels. Let’s get the events for the phantom, where each dipole (1-32) gets its own event:
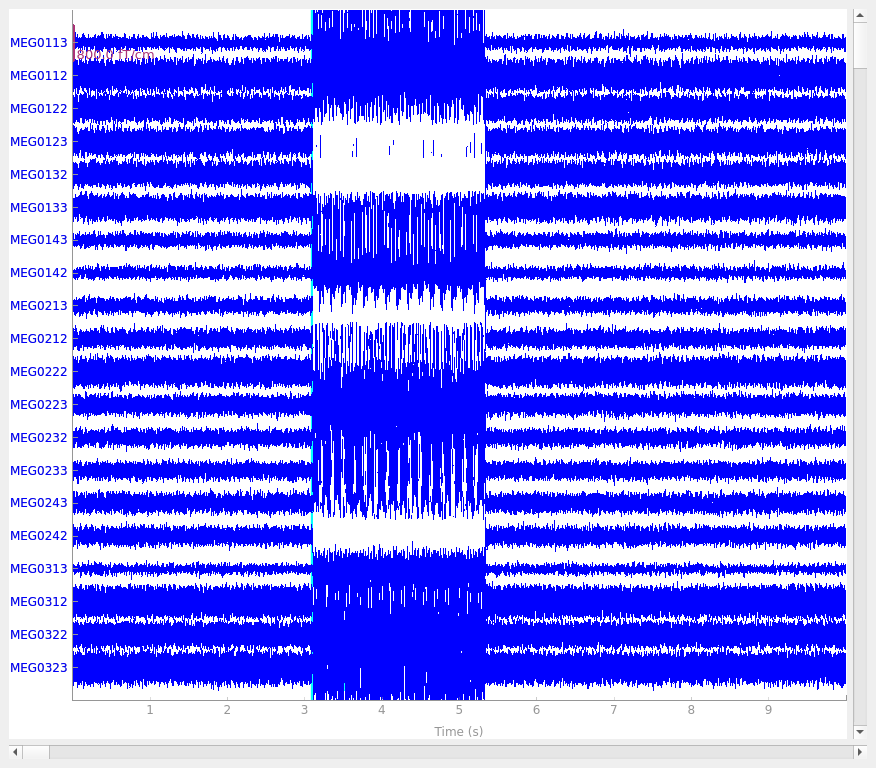
645 events found on stim channel STI201
Event IDs: [ 1 2 3 4 5 6 7 8 9 10 11 12 13 14
15 16 17 18 19 20 21 22 23 24 25 26 27 28
29 30 31 32 256 768 1792 3840 7936]
The data has strong line frequency (60 Hz and harmonics) and cHPI coil noise (five peaks around 300 Hz). Here, we use only the first 30 seconds to save memory:
raw.compute_psd(tmax=30).plot(average=False, picks="data", exclude="bads")
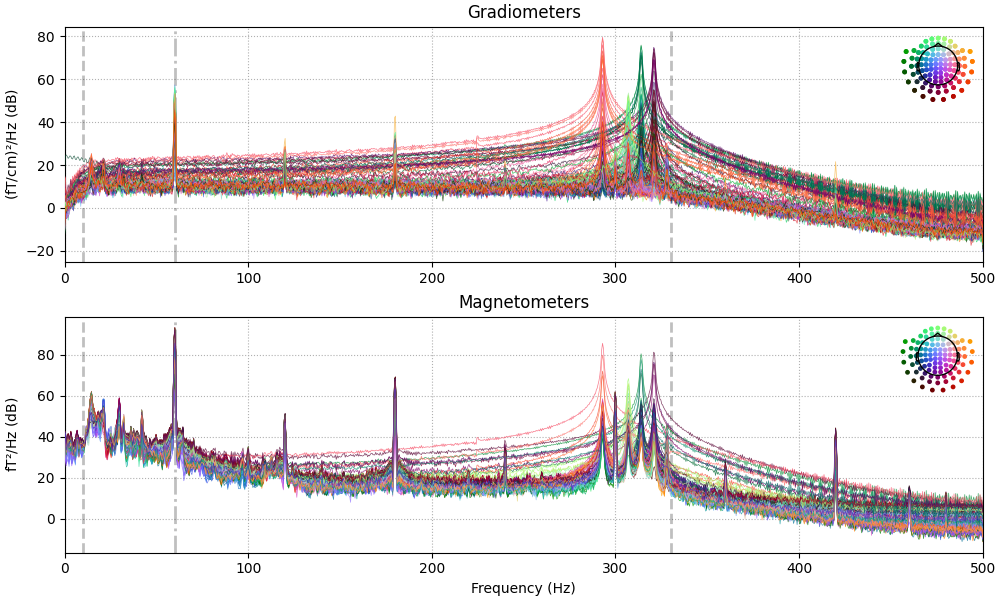
Effective window size : 2.048 (s)
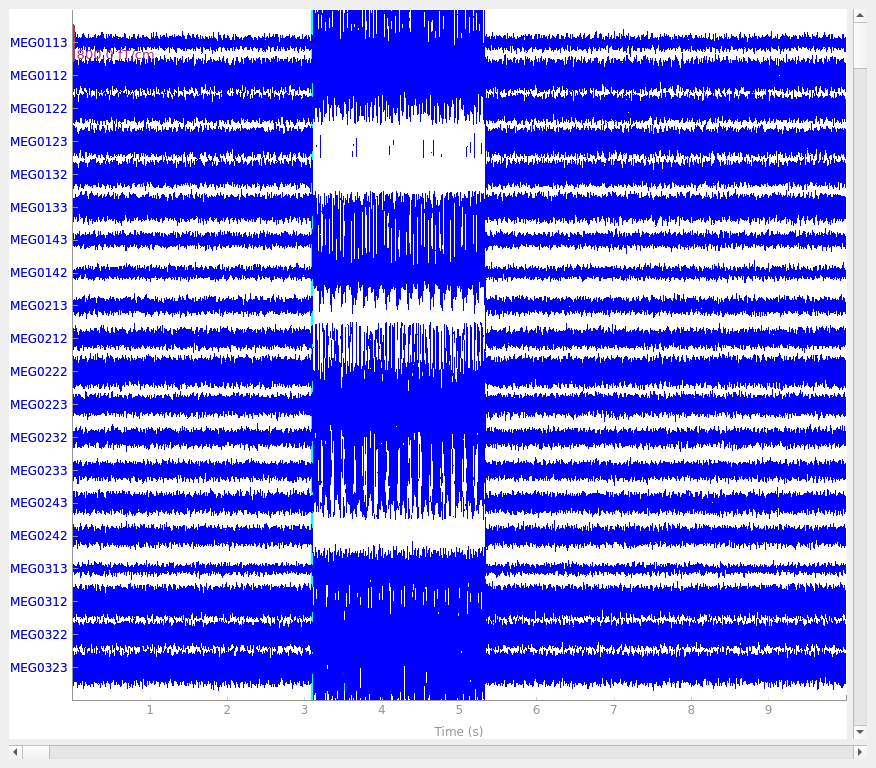
Our phantom produces sinusoidal bursts at 20 Hz:
Now we epoch our data, average it, and look at the first dipole response. The first peak appears around 3 ms. Because we low-passed at 40 Hz, we can also decimate our data to save memory.
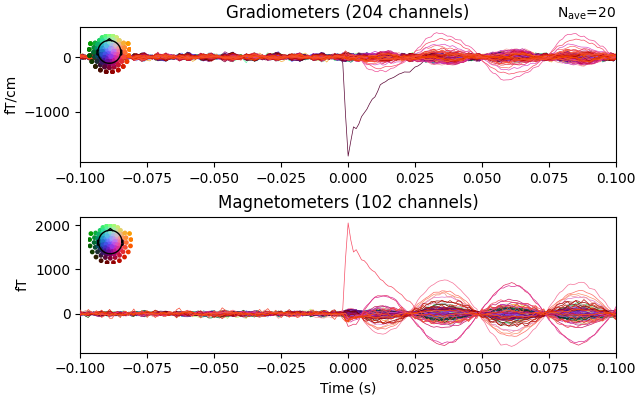
Not setting metadata
640 matching events found
Setting baseline interval to [-0.1, -0.05] s
Applying baseline correction (mode: mean)
Created an SSP operator (subspace dimension = 13)
13 projection items activated
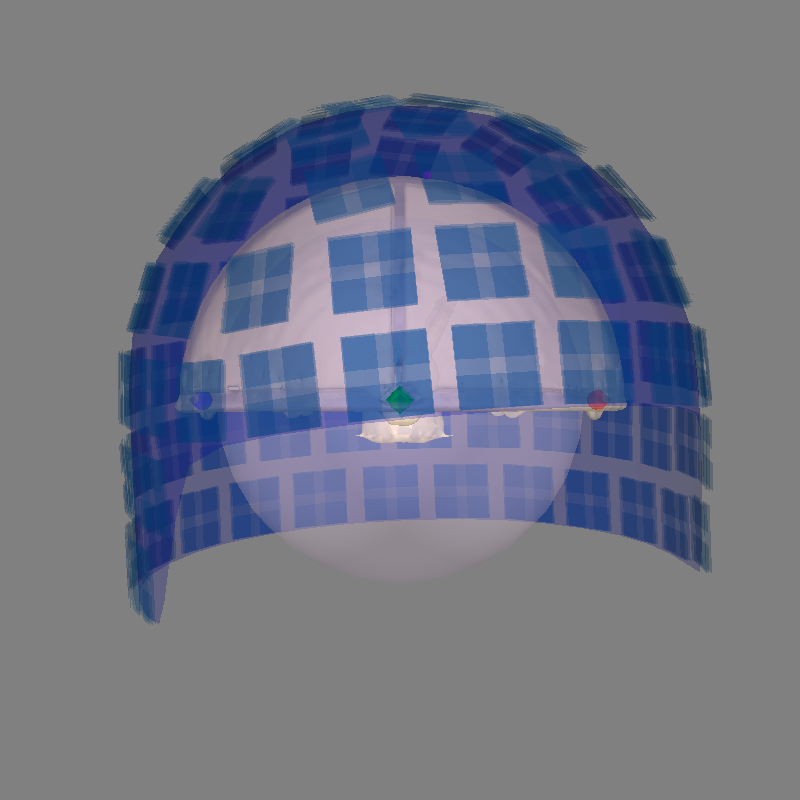
Let’s use a sphere head geometry model and let’s see the coordinate alignment and the sphere location. The phantom is properly modeled by a single-shell sphere with origin (0., 0., 0.).
Even though this is a VectorView/TRIUX phantom, we can use the Otaniemi
phantom subject as a surrogate because the “head” surface (hemisphere outer
shell) has the same geometry for both phantoms, even though the internal
dipole locations differ. The phantom_otaniemi scan was aligned to the
phantom’s head coordinate frame, so an identity trans is appropriate
here.
subjects_dir = data_path
fetch_phantom("otaniemi", subjects_dir=subjects_dir)
sphere = mne.make_sphere_model(r0=(0.0, 0.0, 0.0), head_radius=0.08)
subject = "phantom_otaniemi"
trans = mne.transforms.Transform("head", "mri", np.eye(4))
mne.viz.plot_alignment(
epochs.info,
subject=subject,
show_axes=True,
bem=sphere,
dig=True,
surfaces=("head-dense", "inner_skull"),
trans=trans,
mri_fiducials=True,
subjects_dir=subjects_dir,
)
0 files missing from phantom_otaniemi.txt in /home/circleci/mne_data/MNE-brainstorm-data/bst_phantom_elekta
Equiv. model fitting -> RV = 0.00364477 %
mu1 = 0.944121 lambda1 = 0.138646
mu2 = 0.665982 lambda2 = 0.684475
mu3 = -0.140083 lambda3 = -0.0127517
Set up EEG sphere model with scalp radius 80.0 mm
Using lh.seghead for head surface.
Getting helmet for system 306m
Channel types:: grad: 204, mag: 102
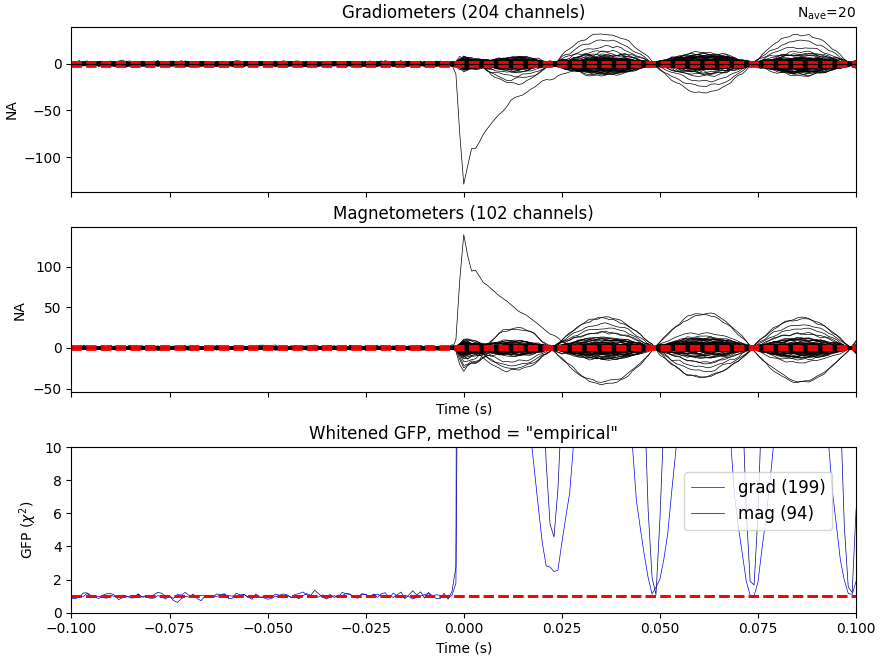
Let’s do some dipole fits. We first compute the noise covariance, then do the fits for each event_id taking the time instant that maximizes the global field power.
# here we can get away with using method='oas' for speed (faster than "shrunk")
# but in general "shrunk" is usually better
cov = mne.compute_covariance(epochs, tmax=bmax)
mne.viz.plot_evoked_white(epochs["1"].average(), cov)
data = []
t_peak = 0.036 # true for Elekta phantom
for ii in event_id:
# Avoid the first and last trials -- can contain dipole-switching artifacts
evoked = epochs[str(ii)][1:-1].average().crop(t_peak, t_peak)
data.append(evoked.data[:, 0])
evoked = mne.EvokedArray(np.array(data).T, evoked.info, tmin=0.0)
del epochs
dip, residual = fit_dipole(evoked, cov, sphere, n_jobs=None)
Loading data for 20 events and 201 original time points ...
0 bad epochs dropped
Loading data for 20 events and 201 original time points ...
0 bad epochs dropped
Loading data for 20 events and 201 original time points ...
0 bad epochs dropped
Loading data for 20 events and 201 original time points ...
0 bad epochs dropped
Loading data for 20 events and 201 original time points ...
0 bad epochs dropped
Loading data for 20 events and 201 original time points ...
0 bad epochs dropped
Loading data for 20 events and 201 original time points ...
0 bad epochs dropped
Loading data for 20 events and 201 original time points ...
0 bad epochs dropped
Loading data for 20 events and 201 original time points ...
0 bad epochs dropped
Loading data for 20 events and 201 original time points ...
0 bad epochs dropped
Loading data for 20 events and 201 original time points ...
0 bad epochs dropped
Loading data for 20 events and 201 original time points ...
0 bad epochs dropped
Loading data for 20 events and 201 original time points ...
0 bad epochs dropped
Loading data for 20 events and 201 original time points ...
0 bad epochs dropped
Loading data for 20 events and 201 original time points ...
0 bad epochs dropped
Loading data for 20 events and 201 original time points ...
0 bad epochs dropped
Loading data for 20 events and 201 original time points ...
0 bad epochs dropped
Loading data for 20 events and 201 original time points ...
0 bad epochs dropped
Loading data for 20 events and 201 original time points ...
0 bad epochs dropped
Loading data for 20 events and 201 original time points ...
0 bad epochs dropped
Loading data for 20 events and 201 original time points ...
0 bad epochs dropped
Loading data for 20 events and 201 original time points ...
0 bad epochs dropped
Loading data for 20 events and 201 original time points ...
0 bad epochs dropped
Loading data for 20 events and 201 original time points ...
0 bad epochs dropped
Loading data for 20 events and 201 original time points ...
0 bad epochs dropped
Loading data for 20 events and 201 original time points ...
0 bad epochs dropped
Loading data for 20 events and 201 original time points ...
0 bad epochs dropped
Loading data for 20 events and 201 original time points ...
0 bad epochs dropped
Loading data for 20 events and 201 original time points ...
0 bad epochs dropped
Loading data for 20 events and 201 original time points ...
0 bad epochs dropped
Loading data for 20 events and 201 original time points ...
0 bad epochs dropped
Loading data for 20 events and 201 original time points ...
0 bad epochs dropped
Computing rank from data with rank=None
Using tolerance 3.9e-08 (2.2e-16 eps * 306 dim * 5.7e+05 max singular value)
Estimated rank (mag + grad): 293
MEG: rank 293 computed from 306 data channels with 13 projectors
Created an SSP operator (subspace dimension = 13)
Setting small MEG eigenvalues to zero (without PCA)
Reducing data rank from 306 -> 293
Estimating covariance using EMPIRICAL
Done.
Number of samples used : 32640
[done]
NOTE: pick_types() is a legacy function. New code should use inst.pick(...).
Computing rank from covariance with rank=None
Using tolerance 3.9e-11 (2.2e-16 eps * 204 dim * 8.5e+02 max singular value)
Estimated rank (grad): 199
GRAD: rank 199 computed from 204 data channels with 5 projectors
Computing rank from covariance with rank=None
Using tolerance 3.5e-14 (2.2e-16 eps * 102 dim * 1.6 max singular value)
Estimated rank (mag): 94
MAG: rank 94 computed from 102 data channels with 8 projectors
Created an SSP operator (subspace dimension = 13)
Computing rank from covariance with rank={'grad': 199, 'mag': 94, 'meg': 293}
Setting small MEG eigenvalues to zero (without PCA)
Created the whitener using a noise covariance matrix with rank 293 (13 small eigenvalues omitted)
BEM : <ConductorModel | Sphere (3 layers): r0=[0.0, 0.0, 0.0] R=80 mm>
MRI transform : identity
Sphere model : origin at ( 0.00 0.00 0.00) mm, rad = 0.1 mm
Guess grid : 20.0 mm
Guess mindist : 5.0 mm
Guess exclude : 20.0 mm
Using normal MEG coil definitions.
Noise covariance : <Covariance | kind : full, shape : (306, 306), range : [-1.8e-21, +2.2e-21], n_samples : 32639>
Coordinate transformation: MRI (surface RAS) -> head
1.000000 0.000000 0.000000 0.00 mm
0.000000 1.000000 0.000000 0.00 mm
0.000000 0.000000 1.000000 0.00 mm
0.000000 0.000000 0.000000 1.00
Coordinate transformation: MEG device -> head
0.976295 -0.211976 0.043756 0.29 mm
0.206488 0.972764 0.105326 0.57 mm
-0.064891 -0.093794 0.993475 5.41 mm
0.000000 0.000000 0.000000 1.00
0 bad channels total
Read 306 MEG channels from info
105 coil definitions read
Coordinate transformation: MEG device -> head
0.976295 -0.211976 0.043756 0.29 mm
0.206488 0.972764 0.105326 0.57 mm
-0.064891 -0.093794 0.993475 5.41 mm
0.000000 0.000000 0.000000 1.00
MEG coil definitions created in head coordinates.
Decomposing the sensor noise covariance matrix...
Created an SSP operator (subspace dimension = 13)
Computing rank from covariance with rank=None
Using tolerance 5.8e-11 (2.2e-16 eps * 306 dim * 8.6e+02 max singular value)
Estimated rank (mag + grad): 293
MEG: rank 293 computed from 306 data channels with 13 projectors
Setting small MEG eigenvalues to zero (without PCA)
Created the whitener using a noise covariance matrix with rank 293 (13 small eigenvalues omitted)
---- Computing the forward solution for the guesses...
Making a spherical guess space with radius 72.0 mm...
Filtering (grid = 20 mm)...
Surface CM = ( 0.0 0.0 0.0) mm
Surface fits inside a sphere with radius 72.0 mm
Surface extent:
x = -72.0 ... 72.0 mm
y = -72.0 ... 72.0 mm
z = -72.0 ... 72.0 mm
Grid extent:
x = -80.0 ... 80.0 mm
y = -80.0 ... 80.0 mm
z = -80.0 ... 80.0 mm
729 sources before omitting any.
178 sources after omitting infeasible sources not within 20.0 - 72.0 mm.
170 sources remaining after excluding the sources outside the surface and less than 5.0 mm inside.
Go through all guess source locations...
[done 170 sources]
---- Fitted : 0.0 ms
---- Fitted : 1.0 ms
---- Fitted : 2.0 ms
---- Fitted : 3.0 ms
---- Fitted : 4.0 ms
---- Fitted : 5.0 ms
---- Fitted : 6.0 ms
---- Fitted : 7.0 ms
---- Fitted : 8.0 ms
---- Fitted : 9.0 ms
---- Fitted : 10.0 ms
---- Fitted : 11.0 ms
---- Fitted : 12.0 ms
---- Fitted : 13.0 ms
---- Fitted : 14.0 ms
---- Fitted : 15.0 ms
---- Fitted : 16.0 ms
[Parallel(n_jobs=1)]: Done 17 tasks | elapsed: 0.7s
---- Fitted : 17.0 ms
---- Fitted : 18.0 ms
---- Fitted : 19.0 ms
---- Fitted : 20.0 ms
---- Fitted : 21.0 ms
---- Fitted : 22.0 ms
---- Fitted : 23.0 ms
---- Fitted : 24.0 ms
---- Fitted : 25.0 ms
---- Fitted : 26.0 ms
---- Fitted : 27.0 ms
---- Fitted : 28.0 ms
---- Fitted : 29.0 ms
---- Fitted : 30.0 ms
---- Fitted : 31.0 ms
Projections have already been applied. Setting proj attribute to True.
32 time points fitted
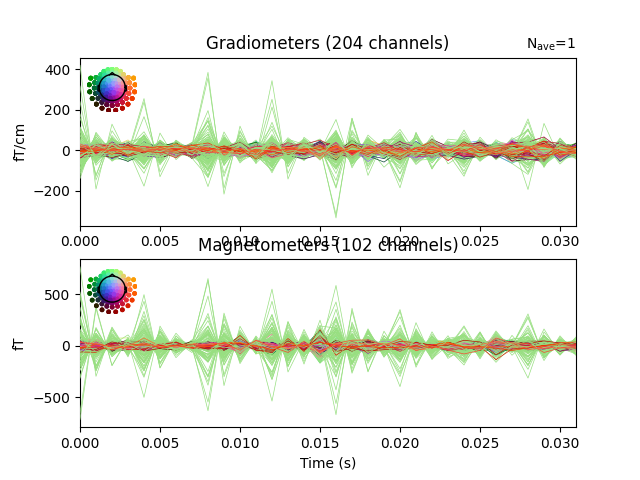
Do a quick visualization of how much variance we explained, putting the data and residuals on the same scale (here the “time points” are the 32 dipole peak values that we fit):
fig, axes = plt.subplots(2, 1)
evoked.plot(axes=axes)
for ax in axes:
for text in list(ax.texts):
text.remove()
for line in ax.lines:
line.set_color("#98df81")
residual.plot(axes=axes)
Now we can compare to the actual locations, taking the difference in mm:
actual_pos, actual_ori = mne.dipole.get_phantom_dipoles()
actual_amp = 100.0 # nAm
fig, (ax1, ax2, ax3) = plt.subplots(
nrows=3, ncols=1, figsize=(6, 7), layout="constrained"
)
diffs = 1000 * np.sqrt(np.sum((dip.pos - actual_pos) ** 2, axis=-1))
print("mean(position error) = %0.1f mm" % (np.mean(diffs),))
ax1.bar(event_id, diffs)
ax1.set_xlabel("Dipole index")
ax1.set_ylabel("Loc. error (mm)")
angles = np.rad2deg(np.arccos(np.abs(np.sum(dip.ori * actual_ori, axis=1))))
print("mean(angle error) = %0.1f°" % (np.mean(angles),))
ax2.bar(event_id, angles)
ax2.set_xlabel("Dipole index")
ax2.set_ylabel("Angle error (°)")
amps = actual_amp - dip.amplitude / 1e-9
print("mean(abs amplitude error) = %0.1f nAm" % (np.mean(np.abs(amps)),))
ax3.bar(event_id, amps)
ax3.set_xlabel("Dipole index")
ax3.set_ylabel("Amplitude error (nAm)")
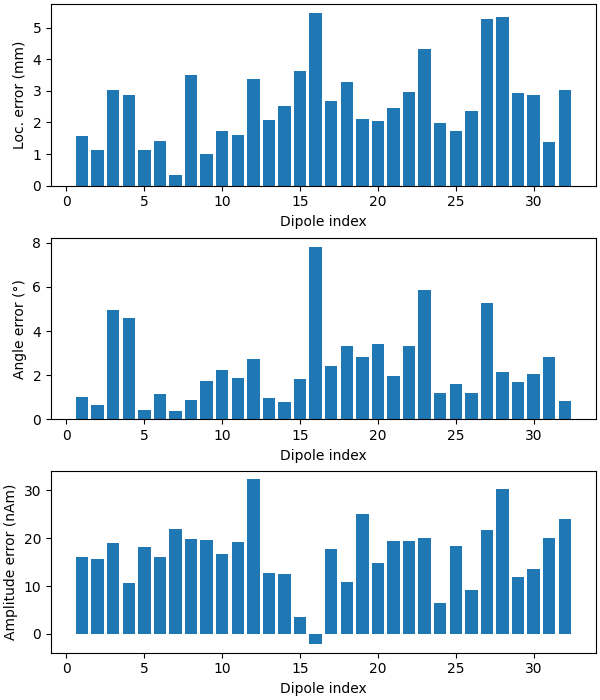
mean(position error) = 2.6 mm
mean(angle error) = 2.4°
mean(abs amplitude error) = 16.8 nAm
Let’s plot the positions and the orientations of the actual and the estimated dipoles
actual_amp = np.ones(len(dip)) # fake amp, needed to create Dipole instance
actual_gof = np.ones(len(dip)) # fake GOF, needed to create Dipole instance
dip_true = mne.Dipole(dip.times, actual_pos, actual_amp, actual_ori, actual_gof)
fig = mne.viz.plot_alignment(
evoked.info,
trans,
subject,
bem=sphere,
surfaces={"head-dense": 0.2},
coord_frame="head",
meg="helmet",
show_axes=True,
subjects_dir=subjects_dir,
)
# Plot the position and the orientation of the actual dipole
fig = mne.viz.plot_dipole_locations(
dipoles=dip_true, mode="arrow", subject=subject, color=(0.0, 0.0, 0.0), fig=fig
)
# Plot the position and the orientation of the estimated dipole
fig = mne.viz.plot_dipole_locations(
dipoles=dip, mode="arrow", subject=subject, color=(0.2, 1.0, 0.5), fig=fig
)
mne.viz.set_3d_view(figure=fig, azimuth=70, elevation=80, distance=0.5)
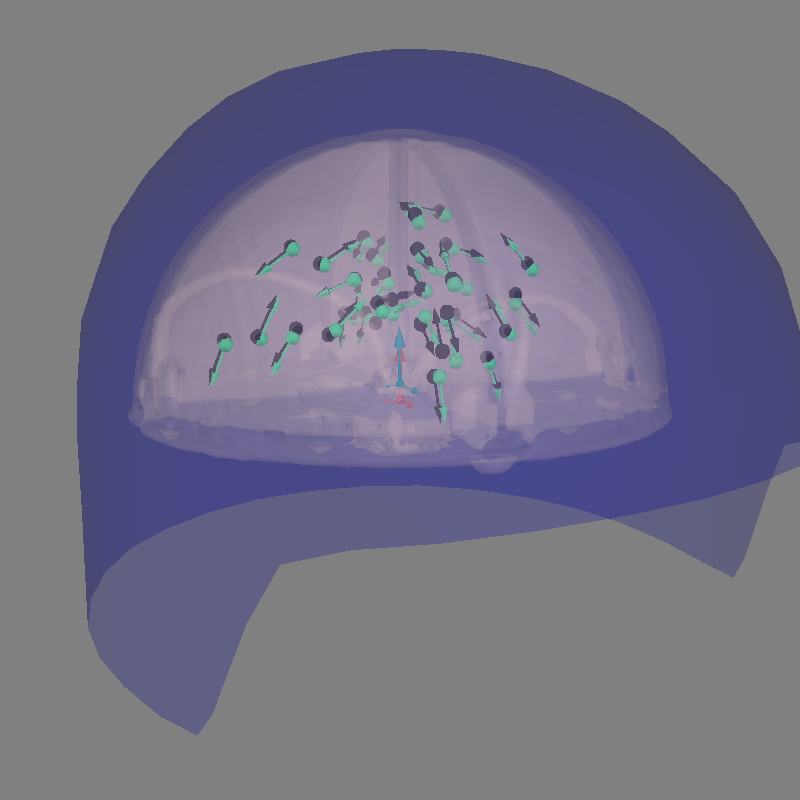
Using lh.seghead for head surface.
Getting helmet for system 306m
References#
Total running time of the script: (0 minutes 49.911 seconds)
Estimated memory usage: 237 MB