Note
Go to the end to download the full example code
4D Neuroimaging/BTi phantom dataset tutorial#
Here we read 4DBTi epochs data obtained with a spherical phantom using four different dipole locations. For each condition we compute evoked data and compute dipole fits.
Data are provided by Jean-Michel Badier from MEG center in Marseille, France.
# Authors: Alex Gramfort <alexandre.gramfort@inria.fr>
#
# License: BSD-3-Clause
# Copyright the MNE-Python contributors.
import os.path as op
import numpy as np
import mne
from mne.datasets import phantom_4dbti
Read data and compute a dipole fit at the peak of the evoked response
data_path = phantom_4dbti.data_path()
raw_fname = op.join(data_path, "%d/e,rfhp1.0Hz")
dipoles = list()
sphere = mne.make_sphere_model(r0=(0.0, 0.0, 0.0), head_radius=0.080)
t0 = 0.07 # peak of the response
pos = np.empty((4, 3))
ori = np.empty((4, 3))
for ii in range(4):
raw = mne.io.read_raw_bti(
raw_fname % (ii + 1,), rename_channels=False, preload=True
)
raw.info["bads"] = ["A173", "A213", "A232"]
events = mne.find_events(raw, "TRIGGER", mask=4350, mask_type="not_and")
epochs = mne.Epochs(
raw, events=events, event_id=8192, tmin=-0.2, tmax=0.4, preload=True
)
evoked = epochs.average()
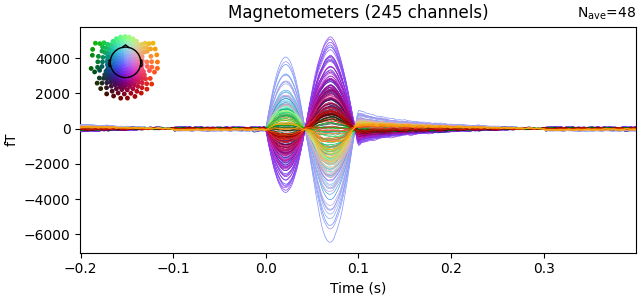
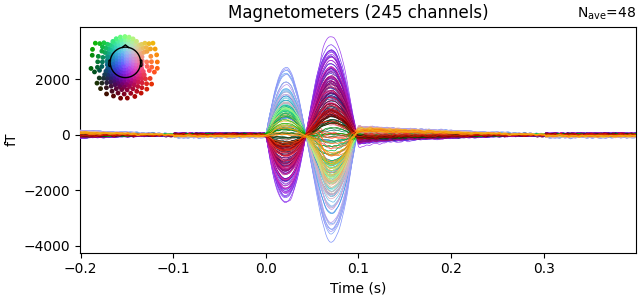
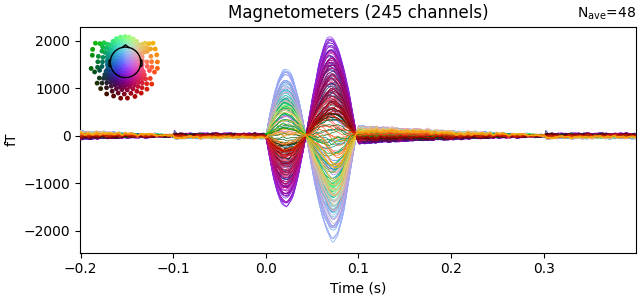
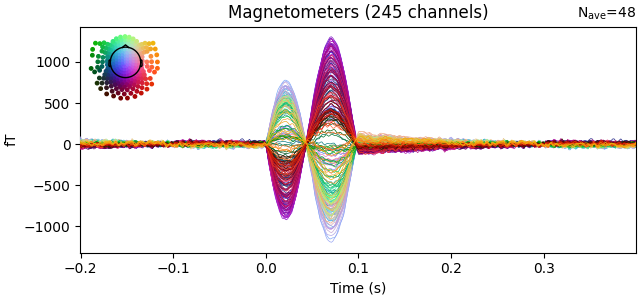
evoked.plot(time_unit="s")
cov = mne.compute_covariance(epochs, tmax=0.0)
dip = mne.fit_dipole(evoked.copy().crop(t0, t0), cov, sphere)[0]
pos[ii] = dip.pos[0]
ori[ii] = dip.ori[0]
Equiv. model fitting -> RV = 0.00364477 %
mu1 = 0.944121 lambda1 = 0.138646
mu2 = 0.665982 lambda2 = 0.684475
mu3 = -0.140083 lambda3 = -0.0127517
Set up EEG sphere model with scalp radius 80.0 mm
Reading 4D PDF file /home/circleci/mne_data/MNE-phantom-4DBTi/1/e,rfhp1.0Hz...
Creating Neuromag info structure ...
... Setting channel info structure.
... putting coil transforms in Neuromag coordinates
... Reading digitization points from /home/circleci/mne_data/MNE-phantom-4DBTi/1/hs_file
Currently direct inclusion of 4D weight tables is not supported. For critical use cases please take into account the MNE command "mne_create_comp_data" to include weights as printed out by the 4D "print_table" routine.
Reading 0 ... 13599 = 0.000 ... 20.052 secs...
Trigger channel TRIGGER has a non-zero initial value of {initial_value} (consider using initial_event=True to detect this event)
50 events found on stim channel TRIGGER
Event IDs: [8192]
Not setting metadata
50 matching events found
Setting baseline interval to [-0.20053968578577042, 0.0] s
Applying baseline correction (mode: mean)
0 projection items activated
Using data from preloaded Raw for 50 events and 408 original time points ...
2 bad epochs dropped
Computing rank from data with rank=None
Using tolerance 3.7e-09 (2.2e-16 eps * 245 dim * 6.9e+04 max singular value)
Estimated rank (mag): 245
MAG: rank 245 computed from 245 data channels with 0 projectors
Reducing data rank from 245 -> 245
Estimating covariance using EMPIRICAL
Done.
Number of samples used : 6576
[done]
BEM : <ConductorModel | Sphere (3 layers): r0=[0.0, 0.0, 0.0] R=80 mm>
MRI transform : identity
Sphere model : origin at ( 0.00 0.00 0.00) mm, rad = 0.1 mm
Guess grid : 20.0 mm
Guess mindist : 5.0 mm
Guess exclude : 20.0 mm
Using normal MEG coil definitions.
Noise covariance : <Covariance | kind : full, shape : (245, 245), range : [-1.1e-26, +4.5e-25], n_samples : 6575>
Coordinate transformation: MRI (surface RAS) -> head
1.000000 0.000000 0.000000 0.00 mm
0.000000 1.000000 0.000000 0.00 mm
0.000000 0.000000 1.000000 0.00 mm
0.000000 0.000000 0.000000 1.00
Coordinate transformation: MEG device -> head
0.975564 -0.033891 -0.217085 -1.50 mm
0.044586 0.998011 0.044560 -34.72 mm
0.215143 -0.053150 0.975135 -6.31 mm
0.000000 0.000000 0.000000 1.00
3 bad channels total
Read 268 MEG channels from info
Read 23 MEG compensation channels from info
0 compensation data sets in info
Setting up compensation data...
No compensation set. Nothing more to do.
105 coil definitions read
Coordinate transformation: MEG device -> head
0.975564 -0.033891 -0.217085 -1.50 mm
0.044586 0.998011 0.044560 -34.72 mm
0.215143 -0.053150 0.975135 -6.31 mm
0.000000 0.000000 0.000000 1.00
MEG coil definitions created in head coordinates.
Decomposing the sensor noise covariance matrix...
Computing rank from covariance with rank=None
Using tolerance 3.9e-14 (2.2e-16 eps * 245 dim * 0.72 max singular value)
Estimated rank (mag): 245
MAG: rank 245 computed from 245 data channels with 0 projectors
Setting small MAG eigenvalues to zero (without PCA)
Created the whitener using a noise covariance matrix with rank 245 (0 small eigenvalues omitted)
---- Computing the forward solution for the guesses...
Making a spherical guess space with radius 72.0 mm...
Filtering (grid = 20 mm)...
Surface CM = ( 0.0 0.0 0.0) mm
Surface fits inside a sphere with radius 72.0 mm
Surface extent:
x = -72.0 ... 72.0 mm
y = -72.0 ... 72.0 mm
z = -72.0 ... 72.0 mm
Grid extent:
x = -80.0 ... 80.0 mm
y = -80.0 ... 80.0 mm
z = -80.0 ... 80.0 mm
729 sources before omitting any.
178 sources after omitting infeasible sources not within 20.0 - 72.0 mm.
170 sources remaining after excluding the sources outside the surface and less than 5.0 mm inside.
Go through all guess source locations...
[done 170 sources]
---- Fitted : 69.3 ms
No projector specified for this dataset. Please consider the method self.add_proj.
1 time points fitted
Reading 4D PDF file /home/circleci/mne_data/MNE-phantom-4DBTi/2/e,rfhp1.0Hz...
Creating Neuromag info structure ...
... Setting channel info structure.
... putting coil transforms in Neuromag coordinates
... Reading digitization points from /home/circleci/mne_data/MNE-phantom-4DBTi/2/hs_file
Currently direct inclusion of 4D weight tables is not supported. For critical use cases please take into account the MNE command "mne_create_comp_data" to include weights as printed out by the 4D "print_table" routine.
Reading 0 ... 13599 = 0.000 ... 20.052 secs...
Trigger channel TRIGGER has a non-zero initial value of {initial_value} (consider using initial_event=True to detect this event)
50 events found on stim channel TRIGGER
Event IDs: [8192]
Not setting metadata
50 matching events found
Setting baseline interval to [-0.20053968578577042, 0.0] s
Applying baseline correction (mode: mean)
0 projection items activated
Using data from preloaded Raw for 50 events and 408 original time points ...
2 bad epochs dropped
Computing rank from data with rank=None
Using tolerance 3.5e-09 (2.2e-16 eps * 245 dim * 6.4e+04 max singular value)
Estimated rank (mag): 245
MAG: rank 245 computed from 245 data channels with 0 projectors
Reducing data rank from 245 -> 245
Estimating covariance using EMPIRICAL
Done.
Number of samples used : 6576
[done]
BEM : <ConductorModel | Sphere (3 layers): r0=[0.0, 0.0, 0.0] R=80 mm>
MRI transform : identity
Sphere model : origin at ( 0.00 0.00 0.00) mm, rad = 0.1 mm
Guess grid : 20.0 mm
Guess mindist : 5.0 mm
Guess exclude : 20.0 mm
Using normal MEG coil definitions.
Noise covariance : <Covariance | kind : full, shape : (245, 245), range : [-2.9e-26, +8.9e-26], n_samples : 6575>
Coordinate transformation: MRI (surface RAS) -> head
1.000000 0.000000 0.000000 0.00 mm
0.000000 1.000000 0.000000 0.00 mm
0.000000 0.000000 1.000000 0.00 mm
0.000000 0.000000 0.000000 1.00
Coordinate transformation: MEG device -> head
0.975554 -0.034041 -0.217109 -1.51 mm
0.044503 0.998063 0.043482 -34.64 mm
0.215208 -0.052081 0.975178 -6.31 mm
0.000000 0.000000 0.000000 1.00
3 bad channels total
Read 268 MEG channels from info
Read 23 MEG compensation channels from info
0 compensation data sets in info
Setting up compensation data...
No compensation set. Nothing more to do.
105 coil definitions read
Coordinate transformation: MEG device -> head
0.975554 -0.034041 -0.217109 -1.51 mm
0.044503 0.998063 0.043482 -34.64 mm
0.215208 -0.052081 0.975178 -6.31 mm
0.000000 0.000000 0.000000 1.00
MEG coil definitions created in head coordinates.
Decomposing the sensor noise covariance matrix...
Computing rank from covariance with rank=None
Using tolerance 3.4e-14 (2.2e-16 eps * 245 dim * 0.63 max singular value)
Estimated rank (mag): 245
MAG: rank 245 computed from 245 data channels with 0 projectors
Setting small MAG eigenvalues to zero (without PCA)
Created the whitener using a noise covariance matrix with rank 245 (0 small eigenvalues omitted)
---- Computing the forward solution for the guesses...
Making a spherical guess space with radius 72.0 mm...
Filtering (grid = 20 mm)...
Surface CM = ( 0.0 0.0 0.0) mm
Surface fits inside a sphere with radius 72.0 mm
Surface extent:
x = -72.0 ... 72.0 mm
y = -72.0 ... 72.0 mm
z = -72.0 ... 72.0 mm
Grid extent:
x = -80.0 ... 80.0 mm
y = -80.0 ... 80.0 mm
z = -80.0 ... 80.0 mm
729 sources before omitting any.
178 sources after omitting infeasible sources not within 20.0 - 72.0 mm.
170 sources remaining after excluding the sources outside the surface and less than 5.0 mm inside.
Go through all guess source locations...
[done 170 sources]
---- Fitted : 69.3 ms
No projector specified for this dataset. Please consider the method self.add_proj.
1 time points fitted
Reading 4D PDF file /home/circleci/mne_data/MNE-phantom-4DBTi/3/e,rfhp1.0Hz...
Creating Neuromag info structure ...
... Setting channel info structure.
... putting coil transforms in Neuromag coordinates
... Reading digitization points from /home/circleci/mne_data/MNE-phantom-4DBTi/3/hs_file
Currently direct inclusion of 4D weight tables is not supported. For critical use cases please take into account the MNE command "mne_create_comp_data" to include weights as printed out by the 4D "print_table" routine.
Reading 0 ... 13599 = 0.000 ... 20.052 secs...
Trigger channel TRIGGER has a non-zero initial value of {initial_value} (consider using initial_event=True to detect this event)
50 events found on stim channel TRIGGER
Event IDs: [8192]
Not setting metadata
50 matching events found
Setting baseline interval to [-0.20053968578577042, 0.0] s
Applying baseline correction (mode: mean)
0 projection items activated
Using data from preloaded Raw for 50 events and 408 original time points ...
2 bad epochs dropped
Computing rank from data with rank=None
Using tolerance 2.6e-09 (2.2e-16 eps * 245 dim * 4.7e+04 max singular value)
Estimated rank (mag): 245
MAG: rank 245 computed from 245 data channels with 0 projectors
Reducing data rank from 245 -> 245
Estimating covariance using EMPIRICAL
Done.
Number of samples used : 6576
[done]
BEM : <ConductorModel | Sphere (3 layers): r0=[0.0, 0.0, 0.0] R=80 mm>
MRI transform : identity
Sphere model : origin at ( 0.00 0.00 0.00) mm, rad = 0.1 mm
Guess grid : 20.0 mm
Guess mindist : 5.0 mm
Guess exclude : 20.0 mm
Using normal MEG coil definitions.
Noise covariance : <Covariance | kind : full, shape : (245, 245), range : [-6.3e-27, +7.5e-26], n_samples : 6575>
Coordinate transformation: MRI (surface RAS) -> head
1.000000 0.000000 0.000000 0.00 mm
0.000000 1.000000 0.000000 0.00 mm
0.000000 0.000000 1.000000 0.00 mm
0.000000 0.000000 0.000000 1.00
Coordinate transformation: MEG device -> head
0.975577 -0.033678 -0.217061 -1.49 mm
0.044611 0.997960 0.045666 -34.78 mm
0.215080 -0.054233 0.975089 -6.31 mm
0.000000 0.000000 0.000000 1.00
3 bad channels total
Read 268 MEG channels from info
Read 23 MEG compensation channels from info
0 compensation data sets in info
Setting up compensation data...
No compensation set. Nothing more to do.
105 coil definitions read
Coordinate transformation: MEG device -> head
0.975577 -0.033678 -0.217061 -1.49 mm
0.044611 0.997960 0.045666 -34.78 mm
0.215080 -0.054233 0.975089 -6.31 mm
0.000000 0.000000 0.000000 1.00
MEG coil definitions created in head coordinates.
Decomposing the sensor noise covariance matrix...
Computing rank from covariance with rank=None
Using tolerance 1.9e-14 (2.2e-16 eps * 245 dim * 0.34 max singular value)
Estimated rank (mag): 245
MAG: rank 245 computed from 245 data channels with 0 projectors
Setting small MAG eigenvalues to zero (without PCA)
Created the whitener using a noise covariance matrix with rank 245 (0 small eigenvalues omitted)
---- Computing the forward solution for the guesses...
Making a spherical guess space with radius 72.0 mm...
Filtering (grid = 20 mm)...
Surface CM = ( 0.0 0.0 0.0) mm
Surface fits inside a sphere with radius 72.0 mm
Surface extent:
x = -72.0 ... 72.0 mm
y = -72.0 ... 72.0 mm
z = -72.0 ... 72.0 mm
Grid extent:
x = -80.0 ... 80.0 mm
y = -80.0 ... 80.0 mm
z = -80.0 ... 80.0 mm
729 sources before omitting any.
178 sources after omitting infeasible sources not within 20.0 - 72.0 mm.
170 sources remaining after excluding the sources outside the surface and less than 5.0 mm inside.
Go through all guess source locations...
[done 170 sources]
---- Fitted : 69.3 ms
No projector specified for this dataset. Please consider the method self.add_proj.
1 time points fitted
Reading 4D PDF file /home/circleci/mne_data/MNE-phantom-4DBTi/4/e,rfhp1.0Hz...
Creating Neuromag info structure ...
... Setting channel info structure.
... putting coil transforms in Neuromag coordinates
... Reading digitization points from /home/circleci/mne_data/MNE-phantom-4DBTi/4/hs_file
Currently direct inclusion of 4D weight tables is not supported. For critical use cases please take into account the MNE command "mne_create_comp_data" to include weights as printed out by the 4D "print_table" routine.
Reading 0 ... 13599 = 0.000 ... 20.052 secs...
Trigger channel TRIGGER has a non-zero initial value of {initial_value} (consider using initial_event=True to detect this event)
50 events found on stim channel TRIGGER
Event IDs: [8192]
Not setting metadata
50 matching events found
Setting baseline interval to [-0.20053968578577042, 0.0] s
Applying baseline correction (mode: mean)
0 projection items activated
Using data from preloaded Raw for 50 events and 408 original time points ...
2 bad epochs dropped
Computing rank from data with rank=None
Using tolerance 2.8e-09 (2.2e-16 eps * 245 dim * 5.1e+04 max singular value)
Estimated rank (mag): 245
MAG: rank 245 computed from 245 data channels with 0 projectors
Reducing data rank from 245 -> 245
Estimating covariance using EMPIRICAL
Done.
Number of samples used : 6576
[done]
BEM : <ConductorModel | Sphere (3 layers): r0=[0.0, 0.0, 0.0] R=80 mm>
MRI transform : identity
Sphere model : origin at ( 0.00 0.00 0.00) mm, rad = 0.1 mm
Guess grid : 20.0 mm
Guess mindist : 5.0 mm
Guess exclude : 20.0 mm
Using normal MEG coil definitions.
Noise covariance : <Covariance | kind : full, shape : (245, 245), range : [-6.4e-27, +3.6e-26], n_samples : 6575>
Coordinate transformation: MRI (surface RAS) -> head
1.000000 0.000000 0.000000 0.00 mm
0.000000 1.000000 0.000000 0.00 mm
0.000000 0.000000 1.000000 0.00 mm
0.000000 0.000000 0.000000 1.00
Coordinate transformation: MEG device -> head
0.975557 -0.033946 -0.217110 -1.50 mm
0.044391 0.998071 0.043409 -34.60 mm
0.215218 -0.051986 0.975181 -6.32 mm
0.000000 0.000000 0.000000 1.00
3 bad channels total
Read 268 MEG channels from info
Read 23 MEG compensation channels from info
0 compensation data sets in info
Setting up compensation data...
No compensation set. Nothing more to do.
105 coil definitions read
Coordinate transformation: MEG device -> head
0.975557 -0.033946 -0.217110 -1.50 mm
0.044391 0.998071 0.043409 -34.60 mm
0.215218 -0.051986 0.975181 -6.32 mm
0.000000 0.000000 0.000000 1.00
MEG coil definitions created in head coordinates.
Decomposing the sensor noise covariance matrix...
Computing rank from covariance with rank=None
Using tolerance 2.1e-14 (2.2e-16 eps * 245 dim * 0.39 max singular value)
Estimated rank (mag): 245
MAG: rank 245 computed from 245 data channels with 0 projectors
Setting small MAG eigenvalues to zero (without PCA)
Created the whitener using a noise covariance matrix with rank 245 (0 small eigenvalues omitted)
---- Computing the forward solution for the guesses...
Making a spherical guess space with radius 72.0 mm...
Filtering (grid = 20 mm)...
Surface CM = ( 0.0 0.0 0.0) mm
Surface fits inside a sphere with radius 72.0 mm
Surface extent:
x = -72.0 ... 72.0 mm
y = -72.0 ... 72.0 mm
z = -72.0 ... 72.0 mm
Grid extent:
x = -80.0 ... 80.0 mm
y = -80.0 ... 80.0 mm
z = -80.0 ... 80.0 mm
729 sources before omitting any.
178 sources after omitting infeasible sources not within 20.0 - 72.0 mm.
170 sources remaining after excluding the sources outside the surface and less than 5.0 mm inside.
Go through all guess source locations...
[done 170 sources]
---- Fitted : 69.3 ms
No projector specified for this dataset. Please consider the method self.add_proj.
1 time points fitted
Compute localisation errors
actual_pos = 0.01 * np.array(
[[0.16, 1.61, 5.13], [0.17, 1.35, 4.15], [0.16, 1.05, 3.19], [0.13, 0.80, 2.26]]
)
actual_pos = np.dot(actual_pos, [[0, 1, 0], [-1, 0, 0], [0, 0, 1]])
errors = 1e3 * np.linalg.norm(actual_pos - pos, axis=1)
print("errors (mm) : %s" % errors)
errors (mm) : [1.37436305 1.3777228 1.21169488 1.17616473]
Plot the dipoles in 3D
actual_amp = np.ones(len(dip)) # fake amp, needed to create Dipole instance
actual_gof = np.ones(len(dip)) # fake GOF, needed to create Dipole instance
dip = mne.Dipole(dip.times, pos, actual_amp, ori, actual_gof)
dip_true = mne.Dipole(dip.times, actual_pos, actual_amp, ori, actual_gof)
fig = mne.viz.plot_alignment(evoked.info, bem=sphere, surfaces=[])
# Plot the position of the actual dipole
fig = mne.viz.plot_dipole_locations(
dipoles=dip_true, mode="sphere", color=(1.0, 0.0, 0.0), fig=fig
)
# Plot the position of the estimated dipole
fig = mne.viz.plot_dipole_locations(
dipoles=dip, mode="sphere", color=(1.0, 1.0, 0.0), fig=fig
)
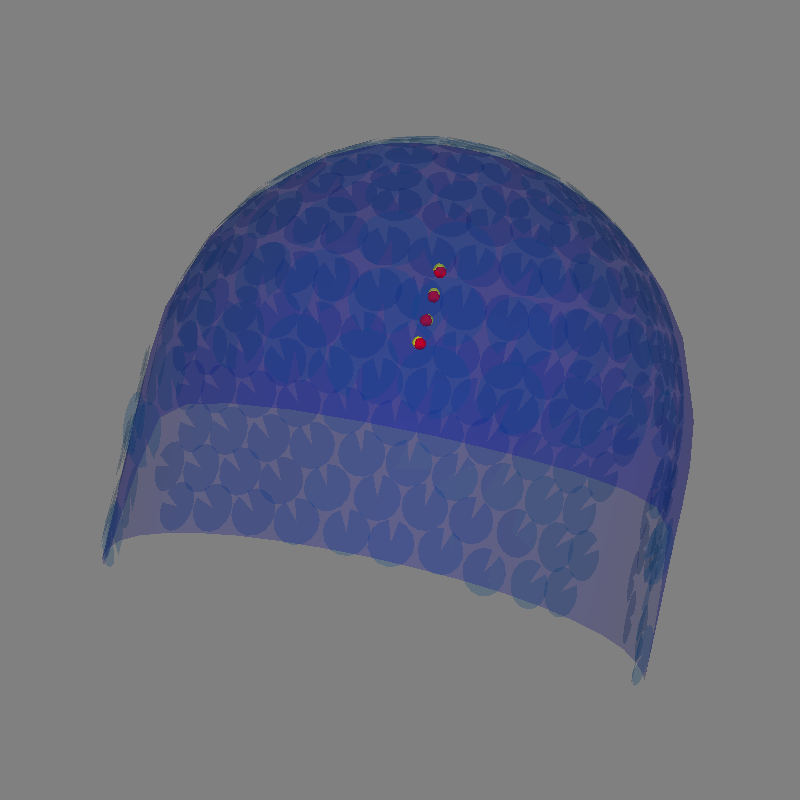
Getting helmet for system Magnes_3600wh
Channel types:: mag: 245
Total running time of the script: (0 minutes 12.483 seconds)
Estimated memory usage: 162 MB