Note
Go to the end to download the full example code
Removing muscle ICA components#
Gross movements produce widespread high-frequency activity across all channels that is usually not recoverable and so the epoch must be rejected as shown in Annotate muscle artifacts. More ubiquitously than gross movements, muscle artifact is produced during postural maintenance. This is more appropriately removed by ICA otherwise there wouldn’t be any epochs left! Note that muscle artifacts of this kind are much more pronounced in EEG than they are in MEG.
# Authors: Alex Rockhill <aprockhill@mailbox.org>
#
# License: BSD-3-Clause
# Copyright the MNE-Python contributors.
import mne
data_path = mne.datasets.sample.data_path()
raw_fname = data_path / "MEG" / "sample" / "sample_audvis_raw.fif"
raw = mne.io.read_raw_fif(raw_fname)
raw.crop(tmin=100, tmax=130) # take 30 seconds for speed
# pick only EEG channels, muscle artifact is basically not picked up by MEG
# if you have a simultaneous recording, you may want to do ICA on MEG and EEG
# separately
raw.pick(picks="eeg", exclude="bads")
# ICA works best with a highpass filter applied
raw.load_data()
raw.filter(l_freq=1.0, h_freq=None)
Opening raw data file /home/circleci/mne_data/MNE-sample-data/MEG/sample/sample_audvis_raw.fif...
Read a total of 3 projection items:
PCA-v1 (1 x 102) idle
PCA-v2 (1 x 102) idle
PCA-v3 (1 x 102) idle
Range : 25800 ... 192599 = 42.956 ... 320.670 secs
Ready.
Removing projector <Projection | PCA-v1, active : False, n_channels : 102>
Removing projector <Projection | PCA-v2, active : False, n_channels : 102>
Removing projector <Projection | PCA-v3, active : False, n_channels : 102>
Reading 0 ... 18019 = 0.000 ... 30.001 secs...
Filtering raw data in 1 contiguous segment
Setting up high-pass filter at 1 Hz
FIR filter parameters
---------------------
Designing a one-pass, zero-phase, non-causal highpass filter:
- Windowed time-domain design (firwin) method
- Hamming window with 0.0194 passband ripple and 53 dB stopband attenuation
- Lower passband edge: 1.00
- Lower transition bandwidth: 1.00 Hz (-6 dB cutoff frequency: 0.50 Hz)
- Filter length: 1983 samples (3.302 s)
[Parallel(n_jobs=1)]: Done 17 tasks | elapsed: 0.0s
Run ICA
ica = mne.preprocessing.ICA(
n_components=15, method="picard", max_iter="auto", random_state=97
)
ica.fit(raw)
Fitting ICA to data using 59 channels (please be patient, this may take a while)
Selecting by number: 15 components
Fitting ICA took 0.6s.
Remove components with postural muscle artifact using ICA
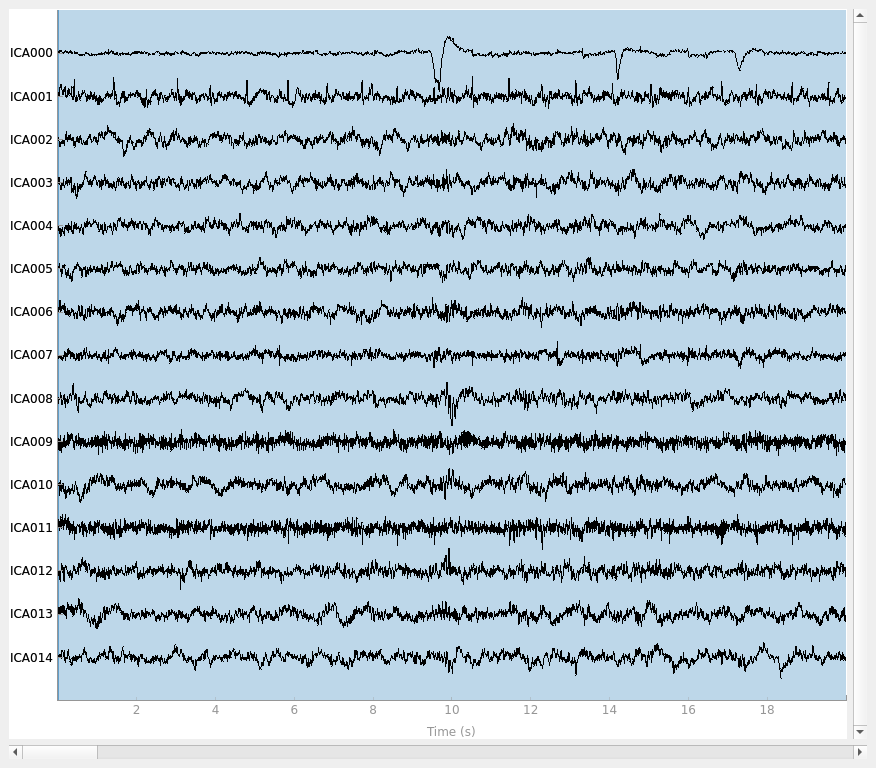
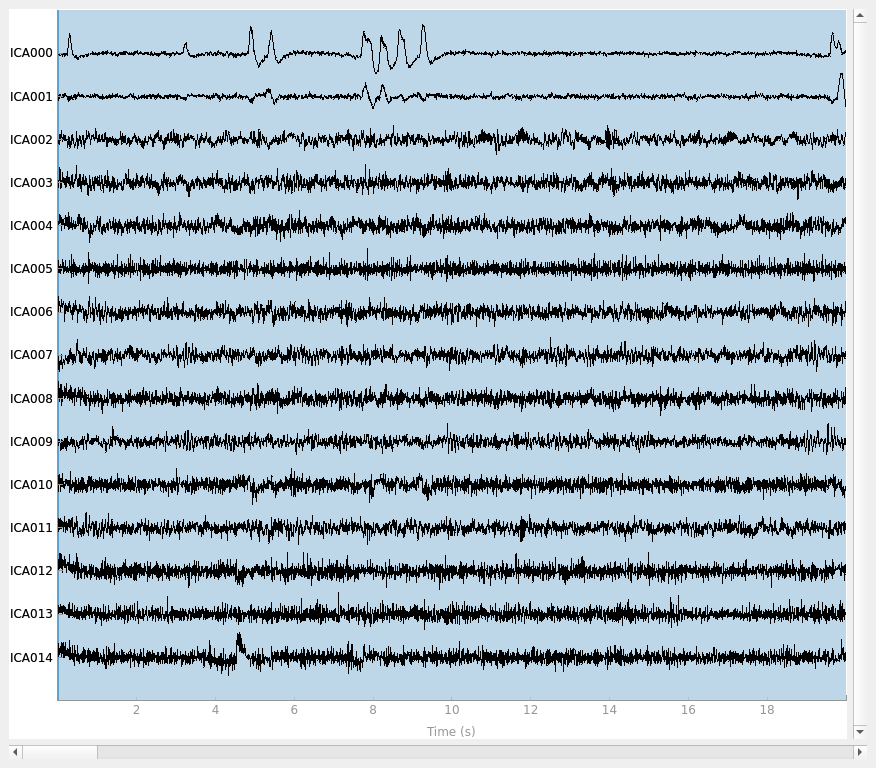
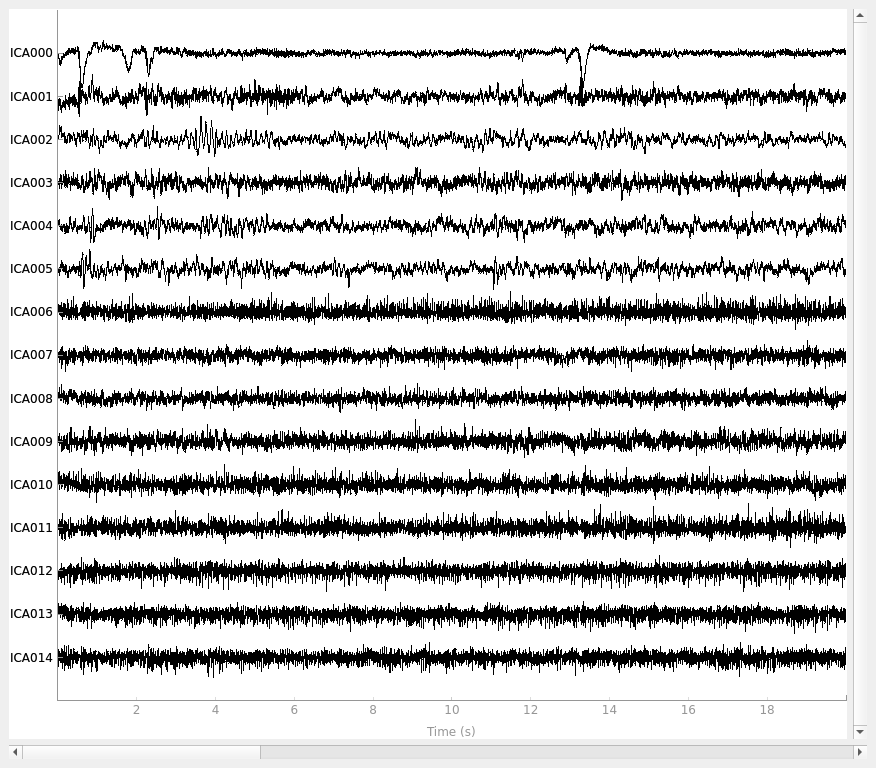
ica.plot_sources(raw)
Creating RawArray with float64 data, n_channels=15, n_times=18020
Range : 85861 ... 103880 = 142.955 ... 172.956 secs
Ready.
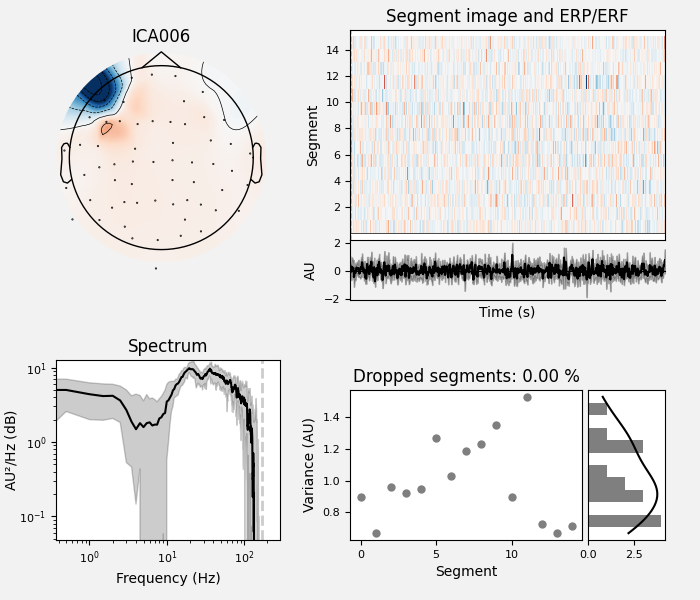
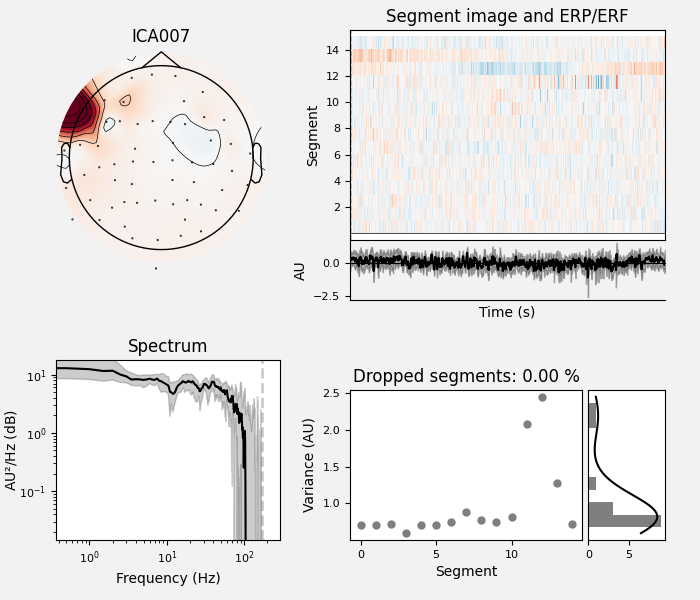
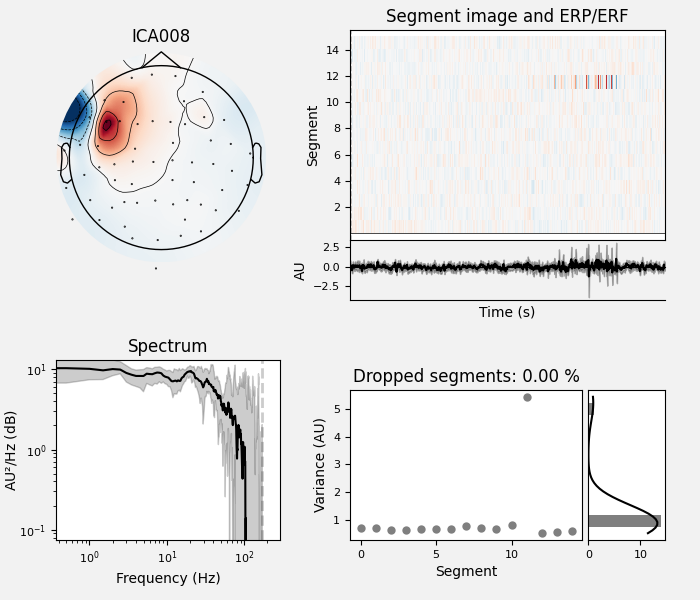
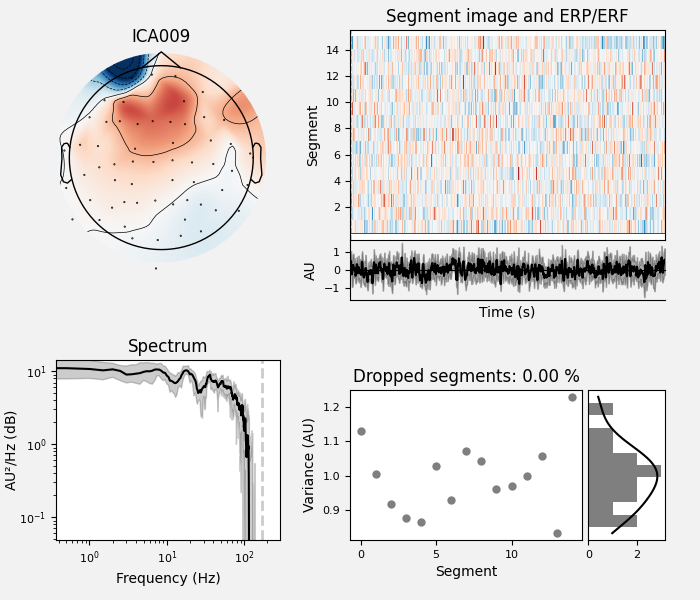
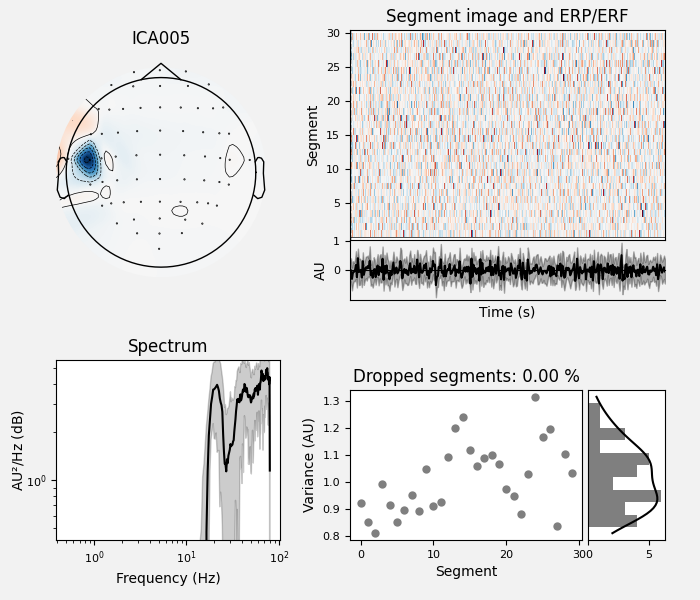
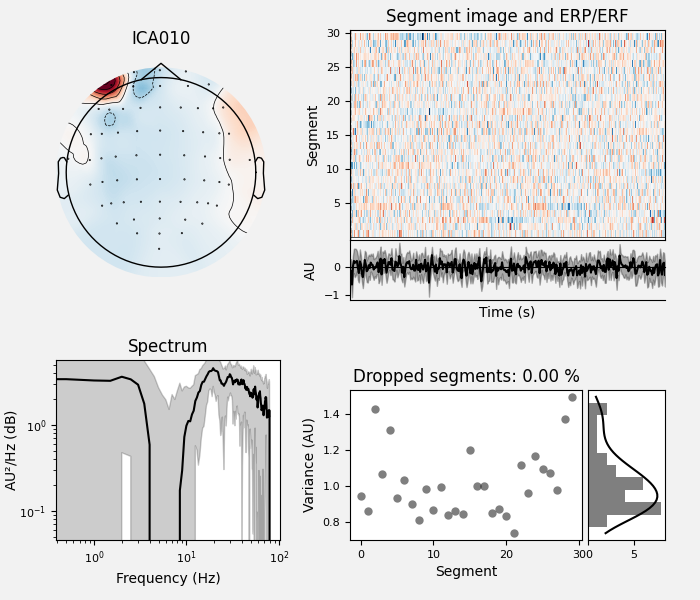
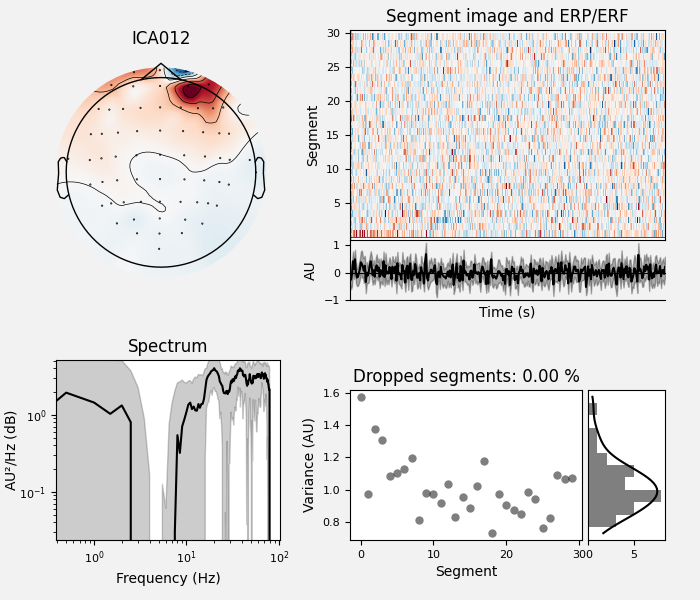
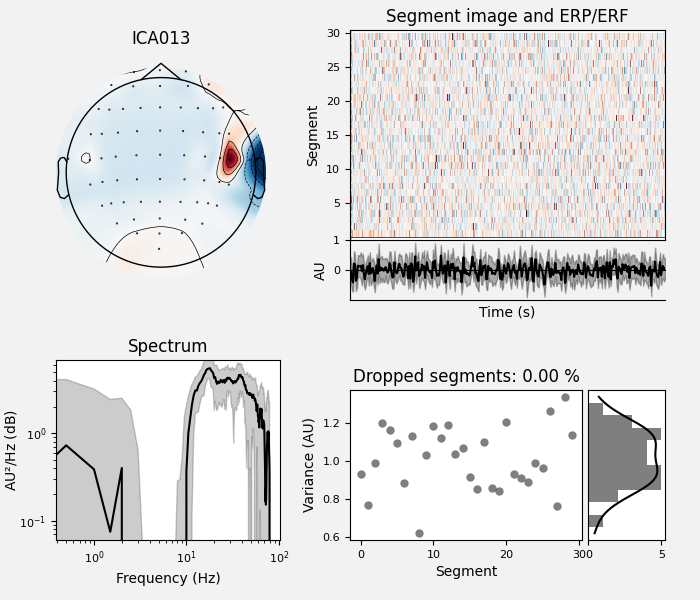
By inspection, let’s select out the muscle-artifact components based on [1] manually.
The criteria are:
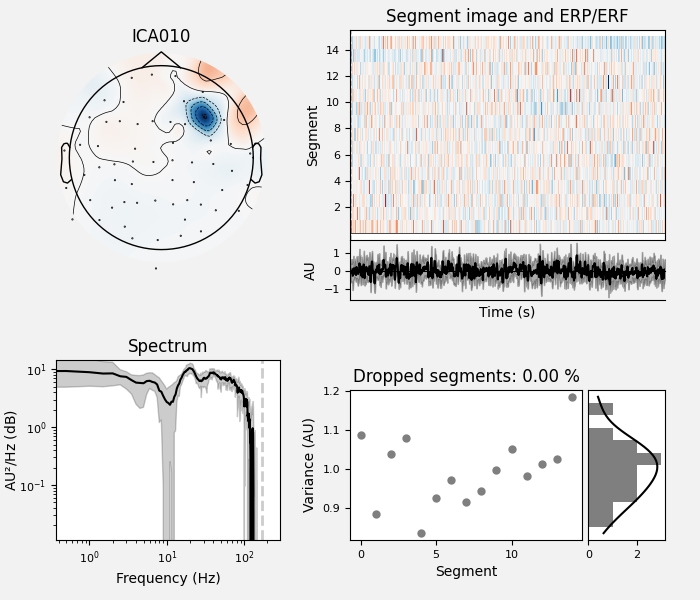
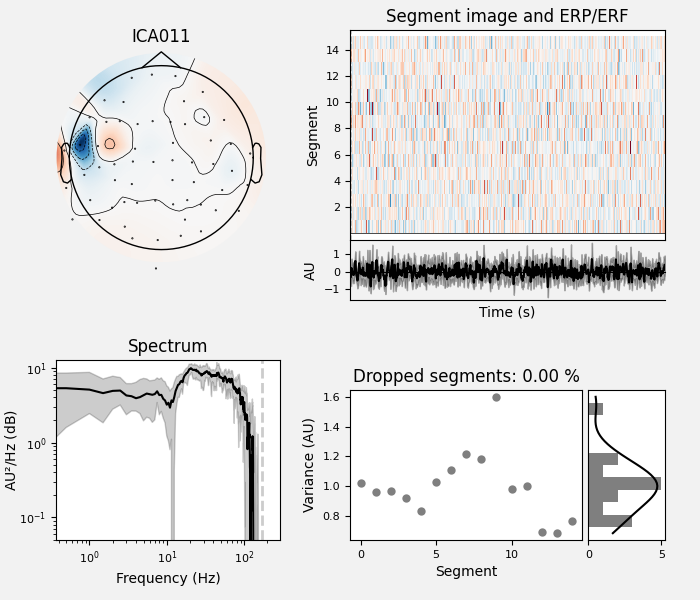
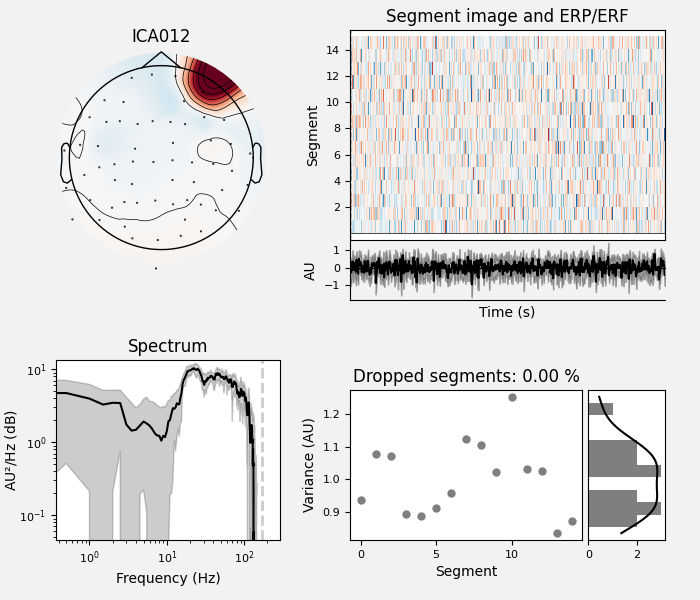
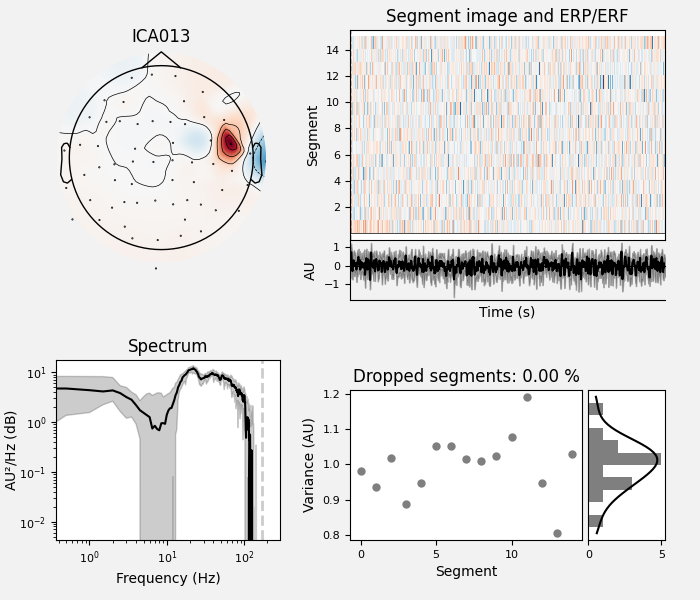
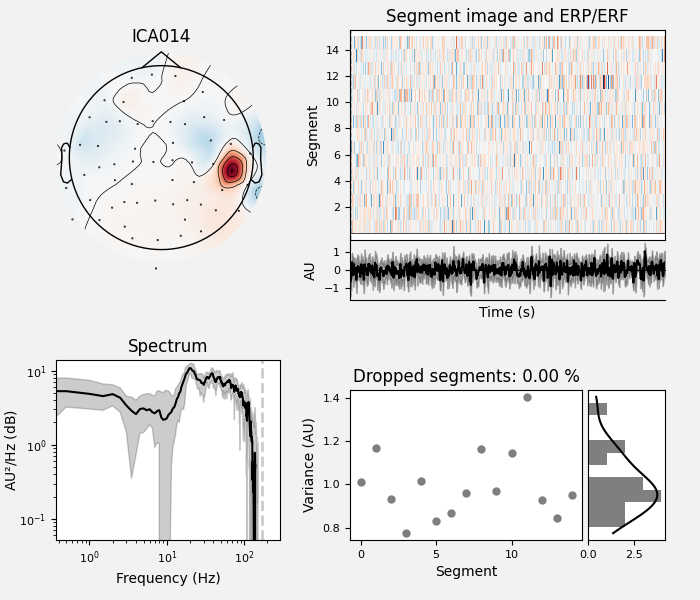
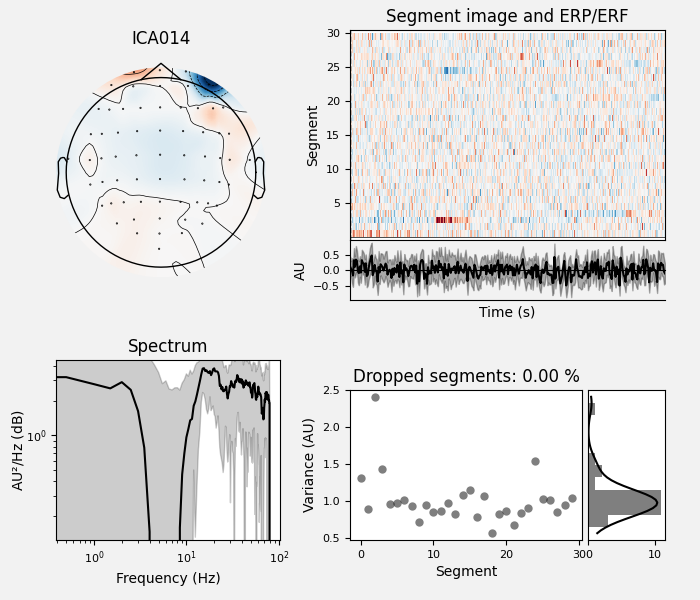
Positive slope of log-log power spectrum between 7 and 75 Hz (here just flat because it’s not in log-log)
Peripheral focus or dipole/multi-pole foci (the blue and red blobs in the topomap are far from the vertex where the most muscle is)
Single focal point (low spatial smoothness; there is just one focus of the topomap compared to components like the first ones that are more likely neural which spread across the topomap)
The other attribute worth noting is that the time course in
mne.preprocessing.ICA.plot_sources() looks like EMG; you can
see spikes when each motor unit fires so that the time course looks fuzzy
and sometimes has large spikes that are often at regular intervals.
ICA component 13 is a textbook example of what muscle artifact looks like. The focus of the topomap for this component is right on the temporalis muscle near the ears. There is also a minimum in the power spectrum at around 10 Hz, then a maximum at around 25 Hz, generally resulting in a positive slope in log-log units; this is a very typical pattern for muscle artifact.
muscle_idx = [6, 7, 8, 9, 10, 11, 12, 13, 14]
ica.plot_properties(raw, picks=muscle_idx, log_scale=True)
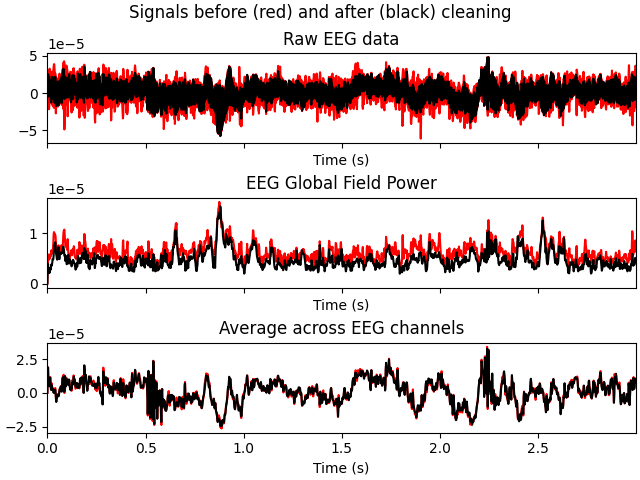
# first, remove blinks and heartbeat to compare
blink_idx = [0]
heartbeat_idx = [5]
ica.apply(raw, exclude=blink_idx + heartbeat_idx)
ica.plot_overlay(raw, exclude=muscle_idx)
Using multitaper spectrum estimation with 7 DPSS windows
Not setting metadata
15 matching events found
No baseline correction applied
0 projection items activated
Not setting metadata
15 matching events found
No baseline correction applied
0 projection items activated
Not setting metadata
15 matching events found
No baseline correction applied
0 projection items activated
Not setting metadata
15 matching events found
No baseline correction applied
0 projection items activated
Not setting metadata
15 matching events found
No baseline correction applied
0 projection items activated
Not setting metadata
15 matching events found
No baseline correction applied
0 projection items activated
Not setting metadata
15 matching events found
No baseline correction applied
0 projection items activated
Not setting metadata
15 matching events found
No baseline correction applied
0 projection items activated
Not setting metadata
15 matching events found
No baseline correction applied
0 projection items activated
Applying ICA to Raw instance
Transforming to ICA space (15 components)
Zeroing out 2 ICA components
Projecting back using 59 PCA components
Applying ICA to Raw instance
Transforming to ICA space (15 components)
Zeroing out 9 ICA components
Projecting back using 59 PCA components
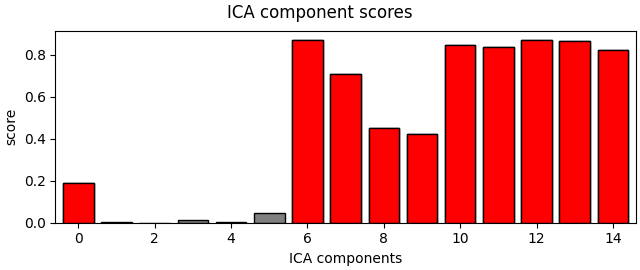
Finally, let’s try an automated algorithm to find muscle components and ensure that it gets the same components we did manually.
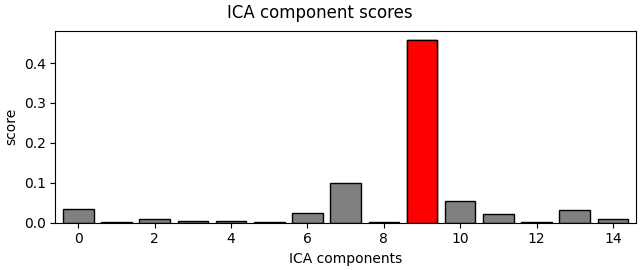
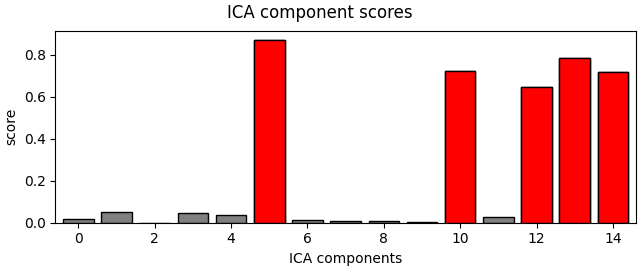
muscle_idx_auto, scores = ica.find_bads_muscle(raw)
ica.plot_scores(scores, exclude=muscle_idx_auto)
print(
f"Manually found muscle artifact ICA components: {muscle_idx}\n"
f"Automatically found muscle artifact ICA components: {muscle_idx_auto}"
)
Effective window size : 3.410 (s)
Manually found muscle artifact ICA components: [6, 7, 8, 9, 10, 11, 12, 13, 14]
Automatically found muscle artifact ICA components: [0, 6, 7, 8, 9, 10, 11, 12, 13, 14]
Let’s now replicate this on the EEGBCI dataset#
for sub in (1, 2):
raw = mne.io.read_raw_edf(
mne.datasets.eegbci.load_data(subject=sub, runs=(1,))[0], preload=True
)
mne.datasets.eegbci.standardize(raw) # set channel names
montage = mne.channels.make_standard_montage("standard_1005")
raw.set_montage(montage)
raw.filter(l_freq=1.0, h_freq=None)
# Run ICA
ica = mne.preprocessing.ICA(
n_components=15, method="picard", max_iter="auto", random_state=97
)
ica.fit(raw)
ica.plot_sources(raw)
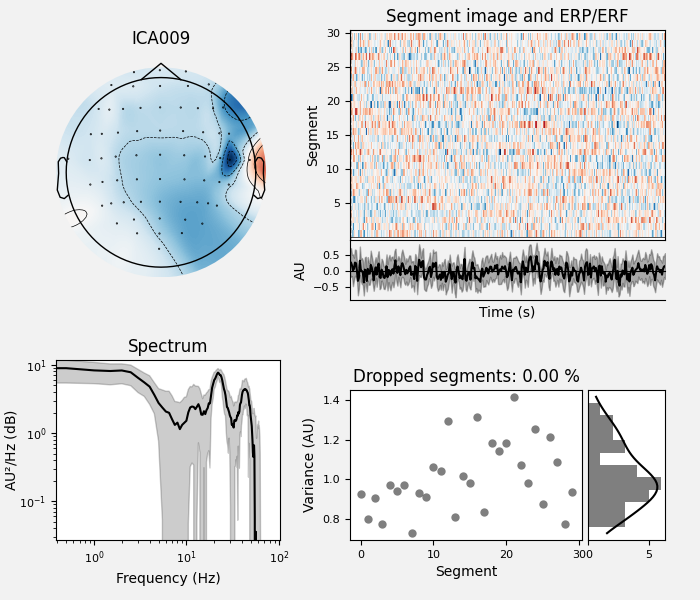
muscle_idx_auto, scores = ica.find_bads_muscle(raw)
ica.plot_properties(raw, picks=muscle_idx_auto, log_scale=True)
ica.plot_scores(scores, exclude=muscle_idx_auto)
print(
f"Manually found muscle artifact ICA components: {muscle_idx}\n"
"Automatically found muscle artifact ICA components: "
f"{muscle_idx_auto}"
)
Extracting EDF parameters from /home/circleci/mne_data/MNE-eegbci-data/files/eegmmidb/1.0.0/S001/S001R01.edf...
EDF file detected
Setting channel info structure...
Creating raw.info structure...
Reading 0 ... 9759 = 0.000 ... 60.994 secs...
Filtering raw data in 1 contiguous segment
Setting up high-pass filter at 1 Hz
FIR filter parameters
---------------------
Designing a one-pass, zero-phase, non-causal highpass filter:
- Windowed time-domain design (firwin) method
- Hamming window with 0.0194 passband ripple and 53 dB stopband attenuation
- Lower passband edge: 1.00
- Lower transition bandwidth: 1.00 Hz (-6 dB cutoff frequency: 0.50 Hz)
- Filter length: 529 samples (3.306 s)
[Parallel(n_jobs=1)]: Done 17 tasks | elapsed: 0.0s
Fitting ICA to data using 64 channels (please be patient, this may take a while)
Selecting by number: 15 components
Fitting ICA took 0.5s.
Creating RawArray with float64 data, n_channels=15, n_times=9760
Range : 0 ... 9759 = 0.000 ... 60.994 secs
Ready.
Effective window size : 12.800 (s)
Using multitaper spectrum estimation with 7 DPSS windows
Not setting metadata
30 matching events found
No baseline correction applied
0 projection items activated
Manually found muscle artifact ICA components: [6, 7, 8, 9, 10, 11, 12, 13, 14]
Automatically found muscle artifact ICA components: [9]
Extracting EDF parameters from /home/circleci/mne_data/MNE-eegbci-data/files/eegmmidb/1.0.0/S002/S002R01.edf...
EDF file detected
Setting channel info structure...
Creating raw.info structure...
Reading 0 ... 9759 = 0.000 ... 60.994 secs...
Filtering raw data in 1 contiguous segment
Setting up high-pass filter at 1 Hz
FIR filter parameters
---------------------
Designing a one-pass, zero-phase, non-causal highpass filter:
- Windowed time-domain design (firwin) method
- Hamming window with 0.0194 passband ripple and 53 dB stopband attenuation
- Lower passband edge: 1.00
- Lower transition bandwidth: 1.00 Hz (-6 dB cutoff frequency: 0.50 Hz)
- Filter length: 529 samples (3.306 s)
[Parallel(n_jobs=1)]: Done 17 tasks | elapsed: 0.0s
Fitting ICA to data using 64 channels (please be patient, this may take a while)
Selecting by number: 15 components
Fitting ICA took 0.8s.
Creating RawArray with float64 data, n_channels=15, n_times=9760
Range : 0 ... 9759 = 0.000 ... 60.994 secs
Ready.
Effective window size : 12.800 (s)
Using multitaper spectrum estimation with 7 DPSS windows
Not setting metadata
30 matching events found
No baseline correction applied
0 projection items activated
Not setting metadata
30 matching events found
No baseline correction applied
0 projection items activated
Not setting metadata
30 matching events found
No baseline correction applied
0 projection items activated
Not setting metadata
30 matching events found
No baseline correction applied
0 projection items activated
Not setting metadata
30 matching events found
No baseline correction applied
0 projection items activated
Manually found muscle artifact ICA components: [6, 7, 8, 9, 10, 11, 12, 13, 14]
Automatically found muscle artifact ICA components: [5, 10, 12, 13, 14]
References#
Total running time of the script: (0 minutes 33.474 seconds)
Estimated memory usage: 13 MB