mne.channels.find_ch_adjacency#
- mne.channels.find_ch_adjacency(info, ch_type)[source]#
Find the adjacency matrix for the given channels.
This function tries to infer the appropriate adjacency matrix template for the given channels. If a template is not found, the adjacency matrix is computed using Delaunay triangulation based on 2D sensor locations.
- Parameters:
- Returns:
- ch_adjacency
scipy.sparse.csr_matrix, shape (n_channels, n_channels) The adjacency matrix.
- ch_names
list The list of channel names present in adjacency matrix.
- ch_adjacency
See also
Notes
New in v0.15.
Automatic detection of an appropriate adjacency matrix template only works for MEG data at the moment. This means that the adjacency matrix is always computed for EEG data and never loaded from a template file. If you want to load a template for a given montage use
read_ch_adjacency()directly.Warning
If Delaunay triangulation is used to calculate the adjacency matrix it may yield partially unexpected results (e.g., include unwanted edges between non-adjacent sensors). Therefore, it is recommended to check (and, if necessary, manually modify) the result by inspecting it via
mne.viz.plot_ch_adjacency().Note that depending on your use case, you may need to additionally use
mne.stats.combine_adjacency()to prepare a final “adjacency” to pass to the eventual function.
Examples using mne.channels.find_ch_adjacency#
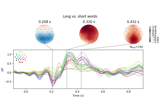
Visualising statistical significance thresholds on EEG data
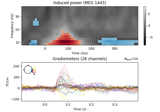
Non-parametric 1 sample cluster statistic on single trial power

Spatiotemporal permutation F-test on full sensor data