Note
Go to the end to download the full example code.
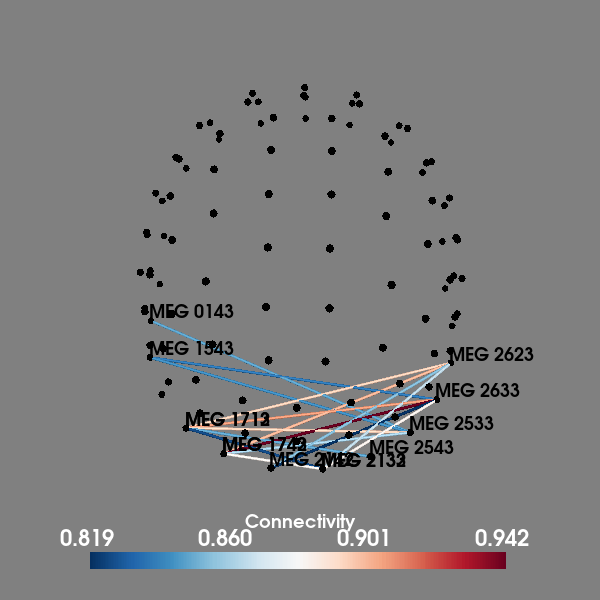
Compute all-to-all connectivity in sensor space#
Computes the Phase Lag Index (PLI) between all gradiometers and shows the connectivity in 3D using the helmet geometry. The left visual stimulation data are used which produces strong connectvitiy in the right occipital sensors.
# Author: Martin Luessi <mluessi@nmr.mgh.harvard.edu>
#
# License: BSD (3-clause)
import os.path as op
import mne
from mne.datasets import sample
from mne_connectivity import spectral_connectivity_epochs
from mne_connectivity.viz import plot_sensors_connectivity
print(__doc__)
# Set parameters
data_path = sample.data_path()
raw_fname = op.join(data_path, "MEG", "sample", "sample_audvis_filt-0-40_raw.fif")
event_fname = op.join(data_path, "MEG", "sample", "sample_audvis_filt-0-40_raw-eve.fif")
# Setup for reading the raw data
raw = mne.io.read_raw_fif(raw_fname)
events = mne.read_events(event_fname)
# Add a bad channel
raw.info["bads"] += ["MEG 2443"]
# Pick MEG gradiometers
picks = mne.pick_types(
raw.info, meg="grad", eeg=False, stim=False, eog=True, exclude="bads"
)
# Create epochs for the visual condition
event_id, tmin, tmax = 3, -0.2, 1.5 # want a long enough epoch for 5 cycles
epochs = mne.Epochs(
raw,
events,
event_id,
tmin,
tmax,
picks=picks,
baseline=(None, 0),
reject=dict(grad=4000e-13, eog=150e-6),
)
epochs.load_data().pick("grad") # just keep MEG and no EOG now
# Compute Fourier coefficients for the epochs (returns an EpochsSpectrum object)
# (storing Fourier coefficients in EpochsSpectrum objects requires MNE >= 1.8)
tmin = 0.0 # exclude the baseline period
spectrum = epochs.compute_psd(method="multitaper", tmin=tmin, output="complex")
# Compute connectivity for the frequency band containing the evoked response
# (passing EpochsSpectrum objects as data requires MNE-Connectivity >= 0.8)
fmin, fmax = 4.0, 9.0
con = spectral_connectivity_epochs(
data=spectrum, method="pli", fmin=fmin, fmax=fmax, faverage=True, n_jobs=1
)
# Now, visualize the connectivity in 3D:
plot_sensors_connectivity(epochs.info, con.get_data(output="dense")[:, :, 0])
Opening raw data file /home/circleci/mne_data/MNE-sample-data/MEG/sample/sample_audvis_filt-0-40_raw.fif...
Read a total of 4 projection items:
PCA-v1 (1 x 102) idle
PCA-v2 (1 x 102) idle
PCA-v3 (1 x 102) idle
Average EEG reference (1 x 60) idle
Range : 6450 ... 48149 = 42.956 ... 320.665 secs
Ready.
Not setting metadata
73 matching events found
Setting baseline interval to [-0.19979521315838786, 0.0] s
Applying baseline correction (mode: mean)
0 projection items activated
Loading data for 73 events and 256 original time points ...
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
24 bad epochs dropped
Using multitaper spectrum estimation with 7 DPSS windows
Connectivity computation...
frequencies: 4.7Hz..8.6Hz (7 points)
connectivity scores will be averaged for each band
only using indices for lower-triangular matrix
computing connectivity for 20503 connections
the following metrics will be computed: PLI
computing cross-spectral density for epoch 1
computing cross-spectral density for epoch 2
computing cross-spectral density for epoch 3
computing cross-spectral density for epoch 4
computing cross-spectral density for epoch 5
computing cross-spectral density for epoch 6
computing cross-spectral density for epoch 7
computing cross-spectral density for epoch 8
computing cross-spectral density for epoch 9
computing cross-spectral density for epoch 10
computing cross-spectral density for epoch 11
computing cross-spectral density for epoch 12
computing cross-spectral density for epoch 13
computing cross-spectral density for epoch 14
computing cross-spectral density for epoch 15
computing cross-spectral density for epoch 16
computing cross-spectral density for epoch 17
computing cross-spectral density for epoch 18
computing cross-spectral density for epoch 19
computing cross-spectral density for epoch 20
computing cross-spectral density for epoch 21
computing cross-spectral density for epoch 22
computing cross-spectral density for epoch 23
computing cross-spectral density for epoch 24
computing cross-spectral density for epoch 25
computing cross-spectral density for epoch 26
computing cross-spectral density for epoch 27
computing cross-spectral density for epoch 28
computing cross-spectral density for epoch 29
computing cross-spectral density for epoch 30
computing cross-spectral density for epoch 31
computing cross-spectral density for epoch 32
computing cross-spectral density for epoch 33
computing cross-spectral density for epoch 34
computing cross-spectral density for epoch 35
computing cross-spectral density for epoch 36
computing cross-spectral density for epoch 37
computing cross-spectral density for epoch 38
computing cross-spectral density for epoch 39
computing cross-spectral density for epoch 40
computing cross-spectral density for epoch 41
computing cross-spectral density for epoch 42
computing cross-spectral density for epoch 43
computing cross-spectral density for epoch 44
computing cross-spectral density for epoch 45
computing cross-spectral density for epoch 46
computing cross-spectral density for epoch 47
computing cross-spectral density for epoch 48
computing cross-spectral density for epoch 49
assembling connectivity matrix
[Connectivity computation done]
<mne.viz.backends._pyvista.PyVistaFigure object at 0x784318328e00>
Total running time of the script: (0 minutes 2.006 seconds)