mne.channels.make_1020_channel_selections#
- mne.channels.make_1020_channel_selections(info, midline='z', *, return_ch_names=False)[source]#
Map hemisphere names to corresponding EEG channel names or indices.
This function uses a simple heuristic to separate channel names into three Region of Interest-based selections:
Left,MidlineandRight.The heuristic is that any of the channel names ending with odd numbers are filed under
Left; those ending with even numbers are filed underRight; and those ending with the character(s) specified inmidlineare filed underMidline. Other channels are ignored.This is appropriate for 10/20, 10/10, 10/05, …, sensor arrangements, but not for other naming conventions.
- Parameters:
- info
mne.Info The
mne.Infoobject with information about the sensors and methods of measurement. If channel locations are present, the channel lists will be sorted from posterior to anterior; otherwise, the order specified ininfo["ch_names"]will be kept.- midline
str Names ending in any of these characters are stored under the
Midlinekey. Defaults to'z'. Capitalization is ignored.- return_ch_names
bool Whether to return channel names instead of channel indices.
New in version 1.4.0.
- info
- Returns:
- selections
dict A dictionary mapping from region of interest name to a list of channel indices (if
return_ch_names=False) or to a list of channel names (ifreturn_ch_names=True).
- selections
Examples using mne.channels.make_1020_channel_selections#
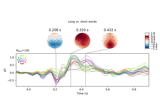
Visualising statistical significance thresholds on EEG data
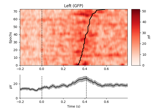
Plot single trial activity, grouped by ROI and sorted by RT