mne.viz.plot_alignment#
- mne.viz.plot_alignment(info=None, trans=None, subject=None, subjects_dir=None, surfaces='auto', coord_frame='auto', meg=None, eeg='original', fwd=None, dig=False, ecog=True, src=None, mri_fiducials=False, bem=None, seeg=True, fnirs=True, show_axes=False, dbs=True, fig=None, interaction='terrain', sensor_colors=None, verbose=None)[source]#
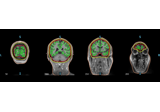
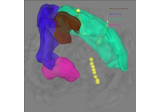
Plot head, sensor, and source space alignment in 3D.
- Parameters:
- info
mne.Info|None The
mne.Infoobject with information about the sensors and methods of measurement. If None (default), no sensor information will be shown.- transpath-like |
dict| instance ofTransform|None If str, the path to the head<->MRI transform
*-trans.fiffile produced during coregistration. Can also be'fsaverage'to use the built-in fsaverage transformation. If trans is None, an identity matrix is assumed. “auto” will load trans from the FreeSurfer directory specified bysubjectandsubjects_dirparameters.Changed in version 0.19: Support for ‘fsaverage’ argument.
- subject
str The FreeSurfer subject name. Can be omitted if
srcis provided.- subjects_dirpath-like |
None The path to the directory containing the FreeSurfer subjects reconstructions. If
None, defaults to theSUBJECTS_DIRenvironment variable.- surfaces
str|list|dict Surfaces to plot. Supported values:
scalp: one of ‘head’, ‘outer_skin’ (alias for ‘head’), ‘head-dense’, or ‘seghead’ (alias for ‘head-dense’)
skull: ‘outer_skull’, ‘inner_skull’, ‘brain’ (alias for ‘inner_skull’)
brain: one of ‘pial’, ‘white’, ‘inflated’, or ‘brain’ (alias for ‘pial’).
Can be dict to specify alpha values for each surface. Use None to specify default value. Specified values must be between 0 and 1. for example:
surfaces=dict(brain=0.4, outer_skull=0.6, head=None)
Defaults to ‘auto’, which will look for a head surface and plot it if found.
Note
For single layer BEMs it is recommended to use
'brain'.- coord_frame‘auto’ | ‘head’ | ‘meg’ | ‘mri’
The coordinate frame to use. If
'auto'(default), chooses'mri'iftranswas passed, and'head'otherwise.Changed in version 1.0: Defaults to
'auto'.- meg
str|list|bool|None Can be “helmet”, “sensors” or “ref” to show the MEG helmet, sensors or reference sensors respectively, or a combination like
('helmet', 'sensors')(same as None, default). True translates to('helmet', 'sensors', 'ref').- eeg
bool|str|list String options are:
- “original” (default; equivalent to
True) Shows EEG sensors using their digitized locations (after transformation to the chosen
coord_frame)
- “original” (default; equivalent to
- “projected”
The EEG locations projected onto the scalp, as is done in forward modeling
Can also be a list of these options, or an empty list (
[], equivalent ofFalse).- fwdinstance of
Forward The forward solution. If present, the orientations of the dipoles present in the forward solution are displayed.
- dig
bool| ‘fiducials’ If True, plot the digitization points; ‘fiducials’ to plot fiducial points only.
- ecog
bool If True (default), show ECoG sensors.
- srcinstance of
SourceSpaces|None If not None, also plot the source space points.
- mri_fiducials
bool|str| path-like Plot MRI fiducials (default False). If
True, look for a file with the canonical name (bem/{subject}-fiducials.fif). Ifstr, it can be'estimated'to usemne.coreg.get_mni_fiducials(), otherwise it should provide the full path to the fiducials file.New in version 0.22: Support for
'estimated'.- bem
listofdict| instance ofConductorModel|None Can be either the BEM surfaces (list of dict), a BEM solution or a sphere model. If None, we first try loading
'$SUBJECTS_DIR/$SUBJECT/bem/$SUBJECT-$SOURCE.fif', and then look for'$SUBJECT*$SOURCE.fif'in the same directory. For'outer_skin', the subjects bem and bem/flash folders are searched. Defaults to None.- seeg
bool If True (default), show sEEG electrodes.
- fnirs
str|list|bool|None Can be “channels”, “pairs”, “detectors”, and/or “sources” to show the fNIRS channel locations, optode locations, or line between source-detector pairs, or a combination like
('pairs', 'channels'). True translates to('pairs',). .. versionadded:: 0.20- show_axes
bool If True (default False), coordinate frame axis indicators will be shown:
head in pink.
MRI in gray (if
trans is not None).MEG in blue (if MEG sensors are present).
New in version 0.16.
- dbs
bool If True (default), show DBS (deep brain stimulation) electrodes.
- fig
Figure3D|None PyVista scene in which to plot the alignment. If
None, creates a new 600x600 pixel figure with black background.New in version 0.16.
- interaction‘trackball’ | ‘terrain’
How interactions with the scene via an input device (e.g., mouse or trackpad) modify the camera position. If
'terrain', one axis is fixed, enabling “turntable-style” rotations. If'trackball', movement along all axes is possible, which provides more freedom of movement, but you may incidentally perform unintentional rotations along some axes.New in version 0.16.
Changed in version 1.0: Defaults to
'terrain'.- sensor_colorsarray_like |
None Colors to use for the sensor glyphs. Can be list-like of color strings (length
n_sensors) or array-like of RGB(A) values (shape(n_sensors, 3)or(n_sensors, 4)).None(the default) uses the default sensor colors for theplot_alignment()GUI.- verbose
bool|str|int|None Control verbosity of the logging output. If
None, use the default verbosity level. See the logging documentation andmne.verbose()for details. Should only be passed as a keyword argument.
- info
- Returns:
- figinstance of
Figure3D The figure.
- figinstance of
See also
Notes
This function serves the purpose of checking the validity of the many different steps of source reconstruction:
Transform matrix (keywords
trans,megandmri_fiducials),BEM surfaces (keywords
bemandsurfaces),sphere conductor model (keywords
bemandsurfaces) andsource space (keywords
surfacesandsrc).
New in version 0.15.
Examples using mne.viz.plot_alignment#
The role of dipole orientations in distributed source localization
EEG source localization given electrode locations on an MRI
Annotate movement artifacts and reestimate dev_head_t
How to convert 3D electrode positions to a 2D image
Compute source power spectral density (PSD) of VectorView and OPM data