mne.minimum_norm.make_inverse_operator#
- mne.minimum_norm.make_inverse_operator(info, forward, noise_cov, loose='auto', depth=0.8, fixed='auto', rank=None, use_cps=True, verbose=None)[source]#
Assemble inverse operator.
- Parameters:
- info
mne.Info The
mne.Infoobject with information about the sensors and methods of measurement. Specifies the channels to include. Bad channels (ininfo['bads']) are not used.- forward
dict Forward operator.
- noise_covinstance of
Covariance The noise covariance matrix.
- loose
float| ‘auto’ |dict Value that weights the source variances of the dipole components that are parallel (tangential) to the cortical surface. Can be:
- float between 0 and 1 (inclusive)
If 0, then the solution is computed with fixed orientation. If 1, it corresponds to free orientations.
'auto'(default)Uses 0.2 for surface source spaces (unless
fixedis True) and 1.0 for other source spaces (volume or mixed).
- dict
Mapping from the key for a given source space type (surface, volume, discrete) to the loose value. Useful mostly for mixed source spaces.
- depth
None|float|dict How to weight (or normalize) the forward using a depth prior. If float (default 0.8), it acts as the depth weighting exponent (
exp) to use None is equivalent to 0, meaning no depth weighting is performed. It can also be adictcontaining keyword arguments to pass tomne.forward.compute_depth_prior()(see docstring for details and defaults). This is effectively ignored whenmethod='eLORETA'.Changed in version 0.20: Depth bias ignored for
method='eLORETA'.- fixed
bool| ‘auto’ Use fixed source orientations normal to the cortical mantle. If True, the loose parameter must be “auto” or 0. If ‘auto’, the loose value is used.
- rank
None| ‘info’ | ‘full’ |dict This controls the rank computation that can be read from the measurement info or estimated from the data. When a noise covariance is used for whitening, this should reflect the rank of that covariance, otherwise amplification of noise components can occur in whitening (e.g., often during source localization).
NoneThe rank will be estimated from the data after proper scaling of different channel types.
'info'The rank is inferred from
info. If data have been processed with Maxwell filtering, the Maxwell filtering header is used. Otherwise, the channel counts themselves are used. In both cases, the number of projectors is subtracted from the (effective) number of channels in the data. For example, if Maxwell filtering reduces the rank to 68, with two projectors the returned value will be 66.'full'The rank is assumed to be full, i.e. equal to the number of good channels. If a
Covarianceis passed, this can make sense if it has been (possibly improperly) regularized without taking into account the true data rank.dictCalculate the rank only for a subset of channel types, and explicitly specify the rank for the remaining channel types. This can be extremely useful if you already know the rank of (part of) your data, for instance in case you have calculated it earlier.
This parameter must be a dictionary whose keys correspond to channel types in the data (e.g.
'meg','mag','grad','eeg'), and whose values are integers representing the respective ranks. For example,{'mag': 90, 'eeg': 45}will assume a rank of90and45for magnetometer data and EEG data, respectively.The ranks for all channel types present in the data, but not specified in the dictionary will be estimated empirically. That is, if you passed a dataset containing magnetometer, gradiometer, and EEG data together with the dictionary from the previous example, only the gradiometer rank would be determined, while the specified magnetometer and EEG ranks would be taken for granted.
The default is
None.- use_cps
bool Whether to use cortical patch statistics to define normal orientations for surfaces (default True).
- verbose
bool|str|int|None Control verbosity of the logging output. If
None, use the default verbosity level. See the logging documentation andmne.verbose()for details. Should only be passed as a keyword argument.
- info
- Returns:
- invinstance of
InverseOperator Inverse operator.
- invinstance of
Notes
For different sets of options (loose, depth, fixed) to work, the forward operator must have been loaded using a certain configuration (i.e., with force_fixed and surf_ori set appropriately). For example, given the desired inverse type (with representative choices of loose = 0.2 and depth = 0.8 shown in the table in various places, as these are the defaults for those parameters):
Inverse desired
Forward parameters allowed
loose
depth
fixed
force_fixed
surf_ori
Loose constraint,Depth weighted0.2
0.8
False
False
True
Loose constraint0.2
None
False
False
True
Free orientation,Depth weighted1.0
0.8
False
False
True
Free orientation1.0
None
False
False
True | False
Fixed constraint,Depth weighted0.0
0.8
True
False
True
Fixed constraint0.0
None
True
True
True
Also note that, if the source space (as stored in the forward solution) has patch statistics computed, these are used to improve the depth weighting. Thus slightly different results are to be expected with and without this information.
For depth weighting, 0.8 is generally good for MEG, and between 2 and 5 is good for EEG, see [1].
References
Examples using mne.minimum_norm.make_inverse_operator#
Working with CTF data: the Brainstorm auditory dataset
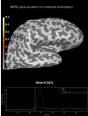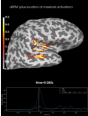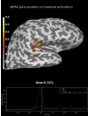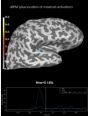
Source localization with MNE, dSPM, sLORETA, and eLORETA
The role of dipole orientations in distributed source localization
EEG source localization given electrode locations on an MRI
Compute source power spectral density (PSD) of VectorView and OPM data
Compute evoked ERS source power using DICS, LCMV beamformer, and dSPM

Compute sparse inverse solution with mixed norm: MxNE and irMxNE
Compute MNE inverse solution on evoked data with a mixed source space
Compute source power estimate by projecting the covariance with MNE

Computing source timecourses with an XFit-like multi-dipole model
Plot point-spread functions (PSFs) and cross-talk functions (CTFs)
Compute spatial resolution metrics in source space
Compute spatial resolution metrics to compare MEG with EEG+MEG