mne.stats.combine_adjacency#
- mne.stats.combine_adjacency(*structure)[source]#
Create a sparse binary adjacency/neighbors matrix.
- Parameters:
- *structure
list The adjacency along each dimension. Each entry can be:
- ndarray or scipy.sparse.spmatrix
A square binary adjacency matrix for the given dimension. For example created by
mne.channels.find_ch_adjacency().
- int
The number of elements along the given dimension. A lattice adjacency will be generated, which is a binary matrix reflecting that element N of an array is adjacent to elements at indices N - 1 and N + 1.
- *structure
- Returns:
- adjacency
scipy.sparse.coo_matrix, shape (n_features, n_features) The square adjacency matrix, where the shape
n_featurescorresponds to the product of the length of all dimensions. For examplelen(times) * len(freqs) * len(chans).
- adjacency
Notes
For 4-dimensional data with shape
(n_obs, n_times, n_freqs, n_chans), you can specify no connections among elements in a particular dimension by passing a matrix of zeros. For example:>>> import numpy as np >>> from scipy.sparse import diags >>> from mne.stats import combine_adjacency >>> n_times, n_freqs, n_chans = (50, 7, 16) >>> chan_adj = diags([1., 1.], offsets=(-1, 1), shape=(n_chans, n_chans)) >>> combine_adjacency( ... n_times, # regular lattice adjacency for times ... np.zeros((n_freqs, n_freqs)), # no adjacency between freq. bins ... chan_adj, # custom matrix, or use mne.channels.find_ch_adjacency ... ) <5600x5600 sparse matrix of type '<class 'numpy.float64'>' with 27076 stored elements in COOrdinate format>
Examples using mne.stats.combine_adjacency#
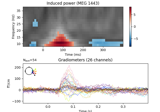
Non-parametric 1 sample cluster statistic on single trial power

Spatiotemporal permutation F-test on full sensor data
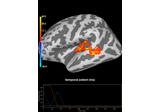
Permutation t-test on source data with spatio-temporal clustering
