Note
Go to the end to download the full example code
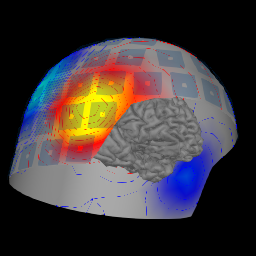
Plot the MNE brain and helmet#
This tutorial shows how to make the MNE helmet + brain image.
Reading inverse operator decomposition from /home/circleci/mne_data/MNE-sample-data/MEG/sample/sample_audvis-meg-oct-6-meg-inv.fif...
Reading inverse operator info...
[done]
Reading inverse operator decomposition...
[done]
305 x 305 full covariance (kind = 1) found.
Read a total of 4 projection items:
PCA-v1 (1 x 102) active
PCA-v2 (1 x 102) active
PCA-v3 (1 x 102) active
Average EEG reference (1 x 60) active
Noise covariance matrix read.
22494 x 22494 diagonal covariance (kind = 2) found.
Source covariance matrix read.
22494 x 22494 diagonal covariance (kind = 6) found.
Orientation priors read.
22494 x 22494 diagonal covariance (kind = 5) found.
Depth priors read.
Did not find the desired covariance matrix (kind = 3)
Reading a source space...
Computing patch statistics...
Patch information added...
Distance information added...
[done]
Reading a source space...
Computing patch statistics...
Patch information added...
Distance information added...
[done]
2 source spaces read
Read a total of 4 projection items:
PCA-v1 (1 x 102) active
PCA-v2 (1 x 102) active
PCA-v3 (1 x 102) active
Average EEG reference (1 x 60) active
Source spaces transformed to the inverse solution coordinate frame
Getting helmet for system 306m
Prepare MEG mapping...
Computing dot products for 305 coils...
[Parallel(n_jobs=1)]: Using backend SequentialBackend with 1 concurrent workers.
[Parallel(n_jobs=1)]: Done 1 out of 1 | elapsed: 1.3s remaining: 0.0s
[Parallel(n_jobs=1)]: Done 1 out of 1 | elapsed: 1.3s finished
Computing dot products for 304 surface locations...
[Parallel(n_jobs=1)]: Using backend SequentialBackend with 1 concurrent workers.
[Parallel(n_jobs=1)]: Done 1 out of 1 | elapsed: 0.8s remaining: 0.0s
[Parallel(n_jobs=1)]: Done 1 out of 1 | elapsed: 0.8s finished
Field mapping data ready
Preparing the mapping matrix...
Truncating at 210/305 components to omit less than 0.0001 (9.9e-05)
Channel types:: grad: 203, mag: 102
import mne
sample_path = mne.datasets.sample.data_path()
subjects_dir = sample_path / "subjects"
fname_evoked = sample_path / "MEG" / "sample" / "sample_audvis-ave.fif"
fname_inv = sample_path / "MEG" / "sample" / "sample_audvis-meg-oct-6-meg-inv.fif"
fname_trans = sample_path / "MEG" / "sample" / "sample_audvis_raw-trans.fif"
inv = mne.minimum_norm.read_inverse_operator(fname_inv)
evoked = mne.read_evokeds(
fname_evoked,
baseline=(None, 0),
proj=True,
verbose=False,
condition="Left Auditory",
)
maps = mne.make_field_map(
evoked,
trans=fname_trans,
ch_type="meg",
subject="sample",
subjects_dir=subjects_dir,
)
time = 0.083
fig = mne.viz.create_3d_figure((256, 256))
mne.viz.plot_alignment(
evoked.info,
subject="sample",
subjects_dir=subjects_dir,
fig=fig,
trans=fname_trans,
meg="sensors",
eeg=False,
surfaces="pial",
coord_frame="mri",
)
evoked.plot_field(maps, time=time, fig=fig, time_label=None, vmax=5e-13)
mne.viz.set_3d_view(
fig,
azimuth=40,
elevation=87,
focalpoint=(0.0, -0.01, 0.04),
roll=-25,
distance=0.55,
)
Total running time of the script: ( 0 minutes 7.069 seconds)
Estimated memory usage: 57 MB