mne_bids.write_anat#
- mne_bids.write_anat(image, bids_path, landmarks=None, deface=False, overwrite=False, verbose=None)[source]#
Put anatomical MRI data into a BIDS format.
Given an MRI scan, format and store the MR data according to BIDS in the correct location inside the specified
mne_bids.BIDSPath. If a transformation matrix is supplied, this information will be stored in a sidecar JSON file.Note
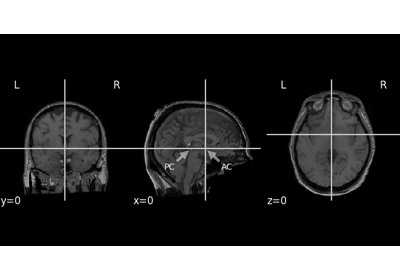
To generate the JSON sidecar with anatomical landmark coordinates (“fiducials”), you need to pass the landmarks via the
landmarksparameter.mne_bids.get_anat_landmarks()may be useful for getting thelandmarks.- Parameters:
- imagepath-like |
NibabelImageObject Path to an MRI scan (e.g. T1w) of the subject. Can be in any format readable by nibabel. Can also be a nibabel image object of an MRI scan. Will be written as a .nii.gz file.
- bids_path
BIDSPath The file to write. The
mne_bids.BIDSPathinstance passed here must have therootandsubjectattributes set. The suffix is assumed to be'T1w'if not present. It can also be'FLASH', for example, to indicate FLASH MRI.- landmarks
mne.channels.DigMontage| path-like |dict|None The montage or path to a montage with landmarks that can be passed to provide information for defacing. Landmarks can be determined from the head model using mne coreg GUI, or they can be determined from the MRI using
freeview. If a dictionary is passed, then the values must be instances ofDigMontageor path-like objects pointing to aDigMontagestored on disk, and the keys of the must be strings (e.g.'session-1') which will be used as naming suffix for the landmarks in the sidecar JSON file. IfNone, no sidecar JSON file will be created.- deface
bool|dict If False, no defacing is performed. If
True, deface with default parameters using the providedlandmarks. If multiple landmarks are provided, will use the ones with the suffix'deface'; if no landmarks with this suffix exist, will use the first ones in thelandmarksdictionary. If dict, accepts the following keys:inset: how far back in voxels to start defacing relative to the nasion (default 5)
theta: is the angle of the defacing shear in degrees relative to vertical (default 15).
- overwrite
bool Whether to overwrite existing files or data in files. Defaults to False. If overwrite is True, any existing files with the same BIDS parameters will be overwritten with the exception of the participants.tsv and scans.tsv files. For these files, parts of pre-existing data that match the current data will be replaced. If overwrite is False, no existing data will be overwritten or replaced.
- verbose
bool|str|int|None Control verbosity of the logging output. If
None, use the default verbosity level. See the logging documentation andmne.verbose()for details. Should only be passed as a keyword argument.
- imagepath-like |
- Returns:
- bids_path
BIDSPath Path to the written MRI data.
- bids_path
Examples using mne_bids.write_anat#
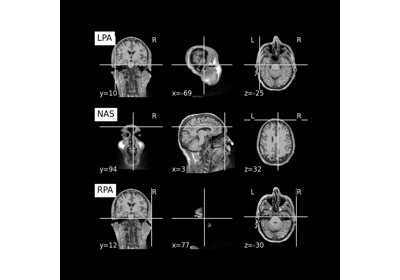
Save and load T1-weighted MRI scan along with anatomical landmarks in BIDS