mne_nirs.preprocessing.quantify_mayer_fooof#
- mne_nirs.preprocessing.quantify_mayer_fooof(raw, num_oscillations=1, centre_frequency=0.01, extra_df_fields=None, fmin=0.001, fmax=1, tmin=0, tmax=None, n_fft=400, n_overlap=200, peak_width_limits=(0.5, 12.0))[source]#
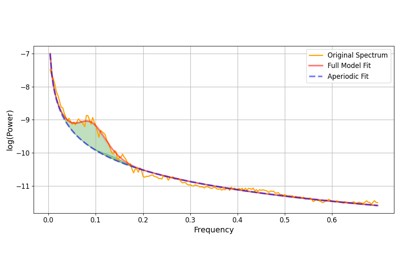
Quantify Mayer wave properties using FOOOF analysis.
The Fitting Oscillations & One Over F (FOOOF) [1] is utilised to estimate Mayer wave oscillation parameters as described in [2].
The FOOOF algorithm is applied to the mean PSD estimate of the data, the oscillation closest to the centre_frequency is assumed to be the Mayer wave oscillation. The parameters for this oscillation are returned as a dataframe. You can return multiple closest oscillations to the centre_frequency by increasing the num_oscillations parameter.
- Parameters:
- rawinstance of
Raw The haemoglobin data.
- num_oscillationsnumber
Number of parameterised oscilations to be returned. These are selected in increasing distance from the centre_frequency.
- centre_frequencynumber
Centre frequency of the Mayer wave.
- extra_df_fieldsnumber
Dictionary of values to be appended to the dataframe.
- fmin
float Min frequency of interest.
- fmax
float Max frequency of interest.
- tmin
float|None Min time of interest.
- tmax
float|None Max time of interest.
- n_fft
int The length of FFT used, must be
>= n_per_seg(default: 256). The segments will be zero-padded ifn_fft > n_per_seg. If n_per_seg is None, n_fft must be <= number of time points in the data.- n_overlap
int The number of points of overlap between segments. Will be adjusted to be <= n_per_seg. The default value is 0.
- peak_width_limits
tupleof (float,float), optional, default: (0.5, 12.0) Limits on possible peak width, in Hz, as (lower_bound, upper_bound). As used by FOOOF.
- rawinstance of
- Returns:
- df
DataFrame Dataframe with columns.
- df
References