Note
Click here to download the full example code
Compute Power Spectral Density of inverse solution from single epochs¶
Compute PSD of dSPM inverse solution on single trial epochs restricted to a brain label. The PSD is computed using a multi-taper method with Discrete Prolate Spheroidal Sequence (DPSS) windows.
# Author: Martin Luessi <mluessi@nmr.mgh.harvard.edu>
#
# License: BSD (3-clause)
import matplotlib.pyplot as plt
import mne
from mne.datasets import sample
from mne.minimum_norm import read_inverse_operator, compute_source_psd_epochs
print(__doc__)
data_path = sample.data_path()
fname_inv = data_path + '/MEG/sample/sample_audvis-meg-oct-6-meg-inv.fif'
fname_raw = data_path + '/MEG/sample/sample_audvis_raw.fif'
fname_event = data_path + '/MEG/sample/sample_audvis_raw-eve.fif'
label_name = 'Aud-lh'
fname_label = data_path + '/MEG/sample/labels/%s.label' % label_name
subjects_dir = data_path + '/subjects'
event_id, tmin, tmax = 1, -0.2, 0.5
snr = 1.0 # use smaller SNR for raw data
lambda2 = 1.0 / snr ** 2
method = "dSPM" # use dSPM method (could also be MNE or sLORETA)
# Load data
inverse_operator = read_inverse_operator(fname_inv)
label = mne.read_label(fname_label)
raw = mne.io.read_raw_fif(fname_raw)
events = mne.read_events(fname_event)
# Set up pick list
include = []
raw.info['bads'] += ['EEG 053'] # bads + 1 more
# pick MEG channels
picks = mne.pick_types(raw.info, meg=True, eeg=False, stim=False, eog=True,
include=include, exclude='bads')
# Read epochs
epochs = mne.Epochs(raw, events, event_id, tmin, tmax, picks=picks,
baseline=(None, 0), reject=dict(mag=4e-12, grad=4000e-13,
eog=150e-6))
# define frequencies of interest
fmin, fmax = 0., 70.
bandwidth = 4. # bandwidth of the windows in Hz
Out:
Reading inverse operator decomposition from /home/circleci/mne_data/MNE-sample-data/MEG/sample/sample_audvis-meg-oct-6-meg-inv.fif...
Reading inverse operator info...
[done]
Reading inverse operator decomposition...
[done]
305 x 305 full covariance (kind = 1) found.
Read a total of 4 projection items:
PCA-v1 (1 x 102) active
PCA-v2 (1 x 102) active
PCA-v3 (1 x 102) active
Average EEG reference (1 x 60) active
Noise covariance matrix read.
22494 x 22494 diagonal covariance (kind = 2) found.
Source covariance matrix read.
22494 x 22494 diagonal covariance (kind = 6) found.
Orientation priors read.
22494 x 22494 diagonal covariance (kind = 5) found.
Depth priors read.
Did not find the desired covariance matrix (kind = 3)
Reading a source space...
Computing patch statistics...
Patch information added...
Distance information added...
[done]
Reading a source space...
Computing patch statistics...
Patch information added...
Distance information added...
[done]
2 source spaces read
Read a total of 4 projection items:
PCA-v1 (1 x 102) active
PCA-v2 (1 x 102) active
PCA-v3 (1 x 102) active
Average EEG reference (1 x 60) active
Source spaces transformed to the inverse solution coordinate frame
Opening raw data file /home/circleci/mne_data/MNE-sample-data/MEG/sample/sample_audvis_raw.fif...
Read a total of 3 projection items:
PCA-v1 (1 x 102) idle
PCA-v2 (1 x 102) idle
PCA-v3 (1 x 102) idle
Range : 25800 ... 192599 = 42.956 ... 320.670 secs
Ready.
Not setting metadata
Not setting metadata
72 matching events found
Setting baseline interval to [-0.19979521315838786, 0.0] sec
Applying baseline correction (mode: mean)
Created an SSP operator (subspace dimension = 3)
3 projection items activated
Compute source space PSD in label¶
- ..note:: By using “return_generator=True” stcs will be a generator object
instead of a list. This allows us so to iterate without having to keep everything in memory.
n_epochs_use = 10
stcs = compute_source_psd_epochs(epochs[:n_epochs_use], inverse_operator,
lambda2=lambda2,
method=method, fmin=fmin, fmax=fmax,
bandwidth=bandwidth, label=label,
return_generator=True, verbose=True)
# compute average PSD over the first 10 epochs
psd_avg = 0.
for i, stc in enumerate(stcs):
psd_avg += stc.data
psd_avg /= n_epochs_use
freqs = stc.times # the frequencies are stored here
stc.data = psd_avg # overwrite the last epoch's data with the average
Out:
Considering frequencies 0 ... 70 Hz
Preparing the inverse operator for use...
Scaled noise and source covariance from nave = 1 to nave = 1
Created the regularized inverter
Created an SSP operator (subspace dimension = 3)
Created the whitener using a noise covariance matrix with rank 302 (3 small eigenvalues omitted)
Computing noise-normalization factors (dSPM)...
[done]
Picked 305 channels from the data
Computing inverse...
Eigenleads need to be weighted ...
Reducing data rank 99 -> 99
Using 2 tapers with bandwidth 4.0 Hz on at most 10 epochs
0%| | : 0/10 [00:00<?, ?it/s]
10%|# | : 1/10 [00:00<00:00, 9.80it/s]
30%|### | : 3/10 [00:00<00:00, 22.91it/s]
40%|#### | : 4/10 [00:00<00:00, 20.13it/s]
50%|##### | : 5/10 [00:00<00:00, 23.54it/s]
70%|####### | : 7/10 [00:00<00:00, 23.96it/s]
90%|######### | : 9/10 [00:00<00:00, 28.67it/s]
100%|##########| : 10/10 [00:00<00:00, 29.41it/s]
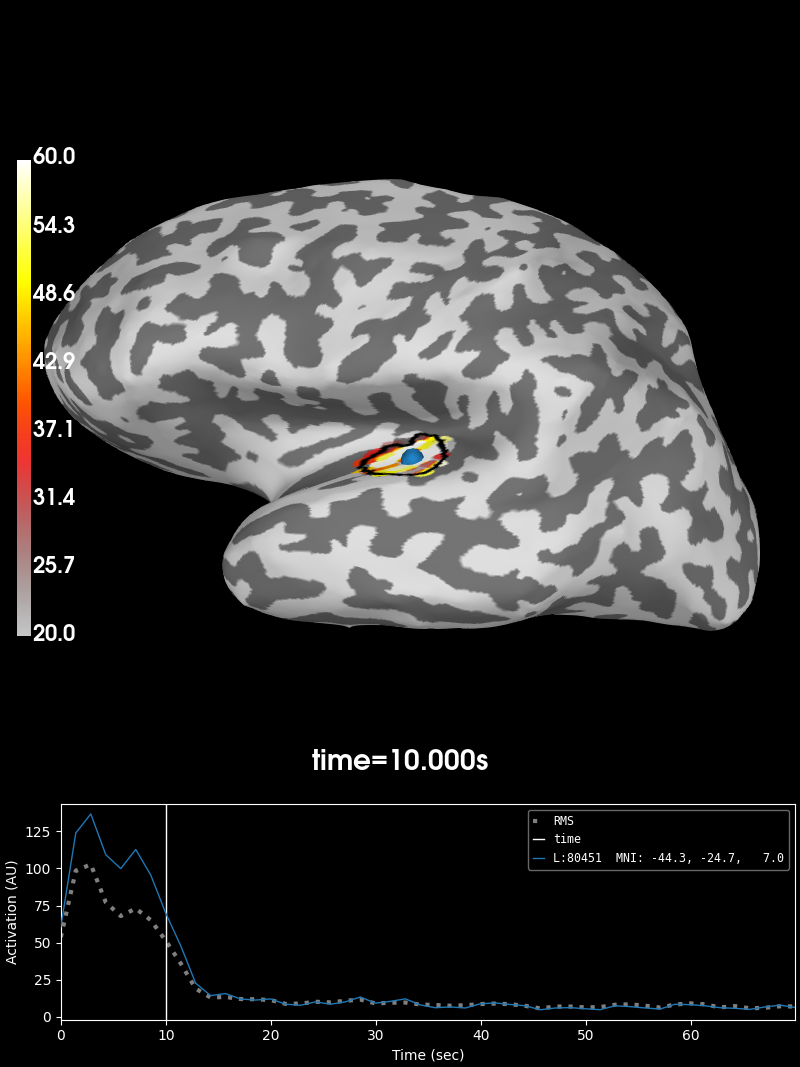
Visualize the 10 Hz PSD:
brain = stc.plot(initial_time=10., hemi='lh', views='lat', # 10 HZ
clim=dict(kind='value', lims=(20, 40, 60)),
smoothing_steps=3, subjects_dir=subjects_dir)
brain.add_label(label, borders=True, color='k')
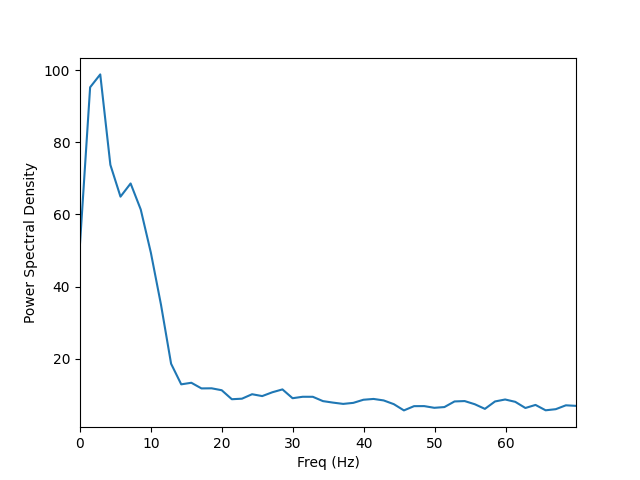
Visualize the entire spectrum:
fig, ax = plt.subplots()
ax.plot(freqs, psd_avg.mean(axis=0))
ax.set_xlabel('Freq (Hz)')
ax.set_xlim(stc.times[[0, -1]])
ax.set_ylabel('Power Spectral Density')
Total running time of the script: ( 0 minutes 10.352 seconds)
Estimated memory usage: 42 MB